Documentation
/ User's manual in C++
: 
Table of Contents
The Modeler module discusses the generation of
surrogate models which aim to provide a simpler, and hence faster, model in order to emulate the specified output of a more
complex model (and generally time and memory consuming) as a function of its inputs and parameters, provided through a TDataServer. The input dataset can either be an existing set of elements
(provided by someone else, resulting from simulations or experiments) or it can be a design-of-experiments generated on purpose, for
the sake of the ongoing study. The meta-model generation is encoded in Uranie with several
different kinds of surrogate model, and also different kinds of possible output format. Once created, the resulting model can
indeed be transmitted to another code and re-used within or without Uranie, in order to avoid regeneration but also
to keep track of achieved performances, as these models can sometimes be created based on a certain randomness (as
discussed in the few sections below).
There are several predefined surrogate-models proposed in the Uranie platform:
The linear regression, discussed in Section V.2
The chaos polynomial expansion, discussed in Section V.3
The artificial neural networks, discussed in Section V.5
The Kriging method, or gaussian process, discussed in Section V.6
It is recommended to follow the law of parsimony (also called Ockham's razor) meaning that the simplest model should be tested first, unless one has insight that it is not well suited for the problem under consideration.
The process of creating a surrogate model within Uranie can be summarised in four simple steps and sketched accordingly as
shown in Figure V.1. Most of these models are also inheriting from the
TModeler class, meaning that they will have common methods and options listed below:
Figure V.1. Simplified decomposition of the model creation process into a four important-step recipe.
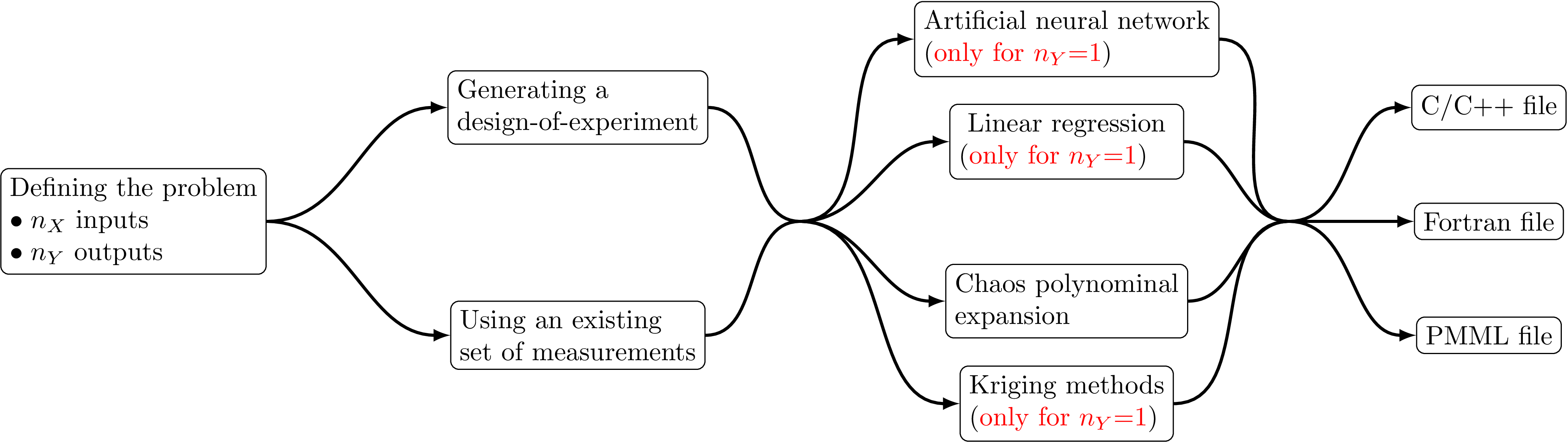 |
As shown in Figure V.1, all these surrogate models need to have a set of elements, that can be a design-of-experiments possibly made specifically for this analysis, or a set of measurements or calculations completely independent of Uranie. This set of elements is used as input to create the model that can then be dumped in different format to be re-used within Uranie or by any other code. It is important to enforce that only the C format can be charge, in ROOT directly via the command:
gROOT->LoadMacro("file_name.C");
The Fortran format cannot be loaded in Uranie, but for some surrogate model it is possible to load the PMML file in the
model and then export it in C file, Uranie can load.
Example:
TANNModeler* tann = new TANNModeler(tds, "rw:r:tu:tl:hu:hl:l:kw,3,yhat");
....
tann->exportFunction("pmml", "file_name.pmml", "function_name","new");
....
TANNModeler* tannPMML = new TANNModeler(tds,"file_name.pmml","function_name");
tannPMML->exportFunction("c++", "new_file_name","function_name");
gROOT->LoadMacro("new_file_name.C"); The table below summarises which type of export can be used with which class of the modeler module.
Table V.1. Type of export allowed for different classes
| c++ | fortran | pmml | |
|---|---|---|---|
TAnisp | X | - | - |
TANNModeler | X | X | X |
TGLM | X | - | - |
TKernel | X | - | - |
TkNN | X | X | - |
TKriging | X | - | X |
TLinearRegression | X | X | X |
TPolynomialChaos | X | - | - |
TPolynomialRegression | X | X | - |