Documentation
/ User's manual in C++
: 
The objective of the macro is to build a multilinear regression between the predictors related to the
normalisation of the variables x1, x2, x3, x4, x5, x6, x7 and a target variable y from the database contained in the ASCII file cornell.dat:
#NAME: cornell
#TITLE: Dataset Cornell 1990
#COLUMN_NAMES: x1 | x2 | x3 | x4 | x5 | x6 | x7 | y
#COLUMN_TITLES: x_{1} | x_{2} | x_{3} | x_{4} | x_{5} | x_{6} | x_{7} | y
0.00 0.23 0.00 0.00 0.00 0.74 0.03 98.7
0.00 0.10 0.00 0.00 0.12 0.74 0.04 97.8
0.00 0.00 0.00 0.10 0.12 0.74 0.04 96.6
0.00 0.49 0.00 0.00 0.12 0.37 0.02 92.0
0.00 0.00 0.00 0.62 0.12 0.18 0.08 86.6
0.00 0.62 0.00 0.00 0.00 0.37 0.01 91.2
0.17 0.27 0.10 0.38 0.00 0.00 0.08 81.9
0.17 0.19 0.10 0.38 0.02 0.06 0.08 83.1
0.17 0.21 0.10 0.38 0.00 0.06 0.08 82.4
0.17 0.15 0.10 0.38 0.02 0.10 0.08 83.2
0.21 0.36 0.12 0.25 0.00 0.00 0.06 81.4
0.00 0.00 0.00 0.55 0.00 0.37 0.08 88.1
{
TDataServer * tds = new TDataServer();
tds->fileDataRead("cornell.dat");
tds->normalize("*", "cr"); // ("x1:x2:x3:x4:x5:x6:y", "cr", TDataServer::kCR)
TCanvas *c = new TCanvas("c1", "Graph for the Macro modeler",5,64,1270,667);
TPad *pad = new TPad("pad","pad",0, 0.03, 1, 1); pad->Draw();
pad->Divide(2);
pad->cd(1);
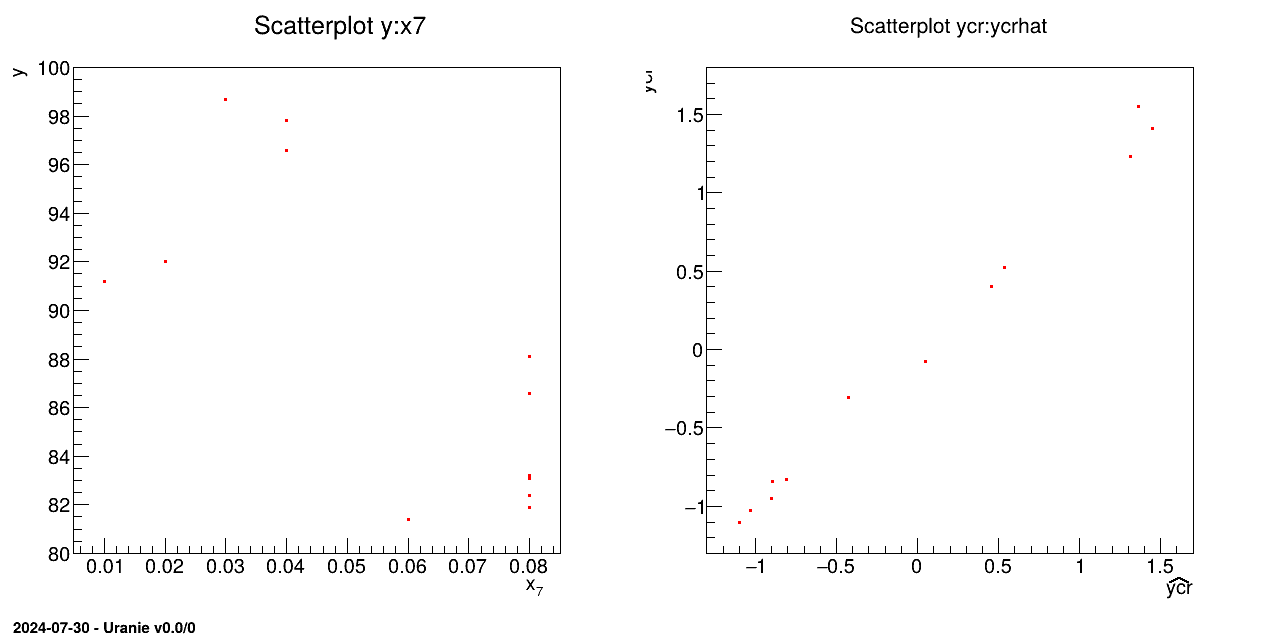
tds->draw("y:x7");
TLinearRegression *tlin = new TLinearRegression(tds,"x1cr:x2cr:x3cr:x4cr:x5cr:x6cr", "ycr", "nointercept");
tlin->estimate();
pad->cd(2);
tds->draw("ycr:ycrhat");
}
The ASCII data file cornell.dat is loaded in the TDataServer:
TDataServer * tds = new TDataServer();
tds->fileDataRead("cornell.dat");The variables are all normalised with a standardised method. The normalised attributes are created with the cr extension:
tds->normalize("*", "cr");
The linear regression is initialised and characteristic values are computed for each normalised variable with the
estimate method. The regression is built with no intercept:
TLinearRegression *tlin = new TLinearRegression(tds,"x1cr:x2cr:x3cr:x4cr:x5cr:x6cr", "ycr", "nointercept");
tlin->estimate();
The objective of this macro is to build a linear regression between a predictor rw and a target variable yhat from the database contained
in the ASCII file flowrate_sampler_launcher_500.dat defining values for the eight variables
described in Section IV.1.2.1 on 500 patterns.
{
TDataServer * tds = new TDataServer();
tds->fileDataRead("flowrate_sampler_launcher_500.dat");
TCanvas *c = new TCanvas("c1", "Graph for the Macro modeler",5,64,1270,667);
TPad *pad = new TPad("pad","pad",0, 0.03, 1, 1); pad->Draw();
pad->Divide(2);
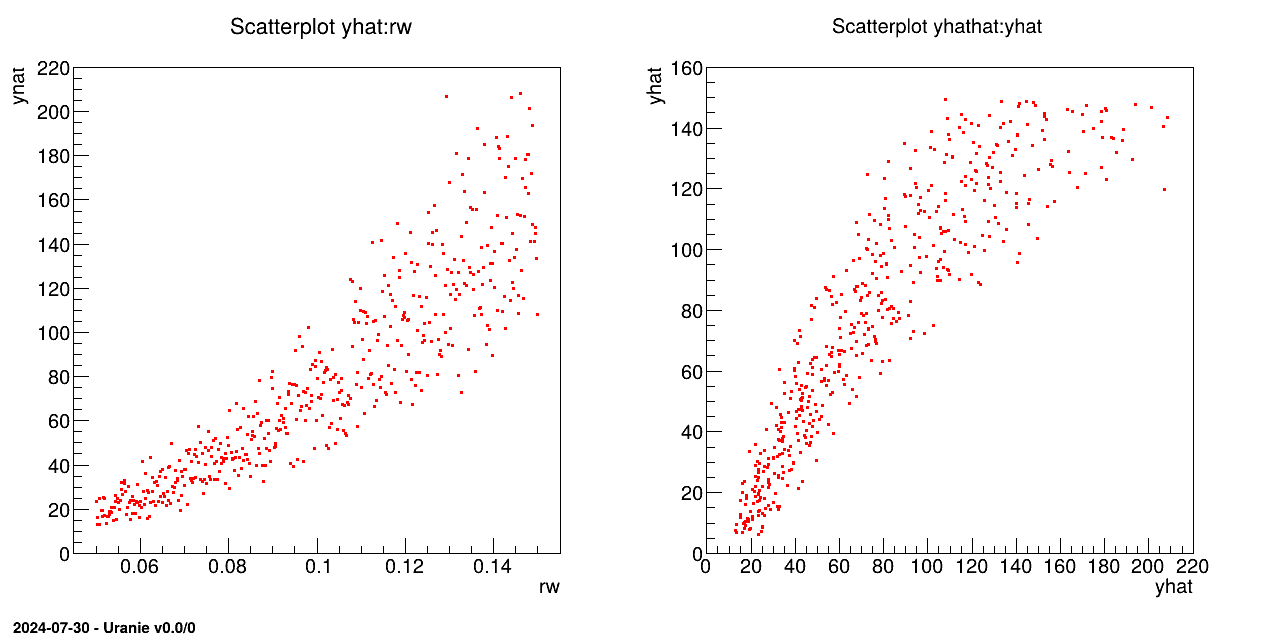
pad->cd(1); tds->draw("yhat:rw");
// tds->getAttribute("yhat")->setOutput();
TLinearRegression *tlin = new TLinearRegression(tds, "rw","yhat");
tlin->estimate();
pad->cd(2); tds->draw("yhathat:yhat");
tds->startViewer();
}
The TDataServer is loaded with the database contained in the file flowrate_sampler_launcher_500.dat
with the fileDataRead method:
TDataServer * tds = new TDataServer();
tds->fileDataRead("flowrate_sampler_launcher_500.dat");
The linear regression is initialised and characteristic values are computed for rw
with the estimate method:
TLinearRegression *tlin = new TLinearRegression(tds, "rw","yhat");
tlin->estimate();
The objective of the macro is to build a multilinear regression between the predictors 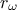 ,
,  ,
, 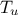 ,
, 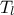 ,
, 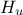 ,
, 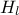 ,
, 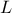 ,
, 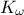 and a target
variable yhat from the database contained in the ASCII file
and a target
variable yhat from the database contained in the ASCII file
flowrate_sampler_launcher_500.dat defining values for these eight variables described in Section IV.1.2.1 on 500 patterns. Parameters of the regression are then
exported in a .C file _FileContainingFunction_.C:
void MultiLinearFunction(double *param, double *res)
{
//////////////////////////////
//
// *********************************************
// ** Uranie v3.12/0 - Date : Thu Jan 04, 2018
// ** Export Modeler : Modeler[LinearRegression]Tds[tdsFlowrate]Predictor[rw:r:tu:tl:hu:hl:l:kw]Target[yhat]
// ** Date : Tue Jan 9 12:08:27 2018
// *********************************************
//
// *********************************************
// ** TDataServer
// ** Name : tdsFlowrate
// ** Title : Design of experiments for Flowrate
// ** Patterns : 500
// ** Attributes : 10
// *********************************************
//
// INPUT : 8
// rw : Min[0.050069828419999997] Max[0.14991758599999999] Unit[]
// r : Min[147.90551809999999] Max[49906.309529999999] Unit[]
// tu : Min[63163.702980000002] Max[115568.17200000001] Unit[]
// tl : Min[63.169232649999998] Max[115.90147020000001] Unit[]
// hu : Min[990.00977509999996] Max[1109.786159] Unit[]
// hl : Min[700.14498509999999] Max[819.81105760000003] Unit[]
// l : Min[1120.3428429999999] Max[1679.3424649999999] Unit[]
// kw : Min[9857.3689890000005] Max[12044.00546] Unit[]
//
// OUTPUT : 1
// yhat : Min[13.09821] Max[208.25110000000001] Unit[]
//
//////////////////////////////
//
// QUALITY :
//
// R2 = 0.948985
// R2a = 0.948154
// Q2 = 0.946835
//
//////////////////////////////
// Intercept
double y = -156.03;
// Attribute : rw
y += param[0]*1422.21;
// Attribute : r
y += param[1]*-3.07412e-07;
// Attribute : tu
y += param[2]*2.15208e-06;
// Attribute : tl
y += param[3]*-0.00498512;
// Attribute : hu
y += param[4]*0.261104;
// Attribute : hl
y += param[5]*-0.254419;
// Attribute : l
y += param[6]*-0.0557145;
// Attribute : kw
y += param[7]*0.00813552;
// Return the value
res[0] = y;
}
This file contains a MultiLinearFunction function. Comparison is made with a database generated from
random variables obeying uniform laws and output variable calculated with this database and the
MultiLinearFunction function.
{
TDataServer * tds = new TDataServer();
tds->fileDataRead("flowrate_sampler_launcher_500.dat");
TCanvas *c = new TCanvas("c1", "Graph for the Macro modeler",5,64,1270,667);
c->Divide(2);
c->cd(1); tds->draw("yhat:rw");
// tds->getAttribute("yhat")->setOutput();
TLinearRegression *tlin = new TLinearRegression(tds, "rw:r:tu:tl:hu:hl:l:kw", "yhat");
tlin->estimate();
tlin->exportFunction("c++", "_FileContainingFunction_", "MultiLinearFunction");
// Define the DataServer
TDataServer *tds2 = new TDataServer("tdsFlowrate", "Design of experiments for Flowrate");
// Add the study attributes
tds2->addAttribute( new TUniformDistribution("rw", 0.05, 0.15));
tds2->addAttribute( new TUniformDistribution("r", 100.0, 50000.0));
tds2->addAttribute( new TUniformDistribution("tu", 63070.0, 115600.0));
tds2->addAttribute( new TUniformDistribution("tl", 63.1, 116.0));
tds2->addAttribute( new TUniformDistribution("hu", 990.0, 1110.0));
tds2->addAttribute( new TUniformDistribution("hl", 700.0, 820.0));
tds2->addAttribute( new TUniformDistribution("l", 1120.0, 1680.0));
tds2->addAttribute( new TUniformDistribution("kw", 9855.0, 12045.0));
Int_t nS = 1000;
// \test Générateur de plan d'expérience
TSampling *sampling = new TSampling(tds2, "lhs", nS);
sampling->generateSample();
gROOT->LoadMacro("_FileContainingFunction_.C");
// Create a TLauncherFunction from a TDataServer and an analytical function
// Rename the outpout attribute "ymod"
TLauncherFunction * tlf = new TLauncherFunction(tds2, "MultiLinearFunction","","ymod");
// Evaluate the function on all the design of experiments
tlf->run();
c->Clear();
tds2->getTuple()->SetMarkerColor(kBlue);
tds2->getTuple()->SetMarkerStyle(8);
tds2->getTuple()->SetMarkerSize(1.2);
tds->getTuple()->SetMarkerColor(kGreen);
tds->getTuple()->SetMarkerStyle(8);
tds->getTuple()->SetMarkerSize(1.2);
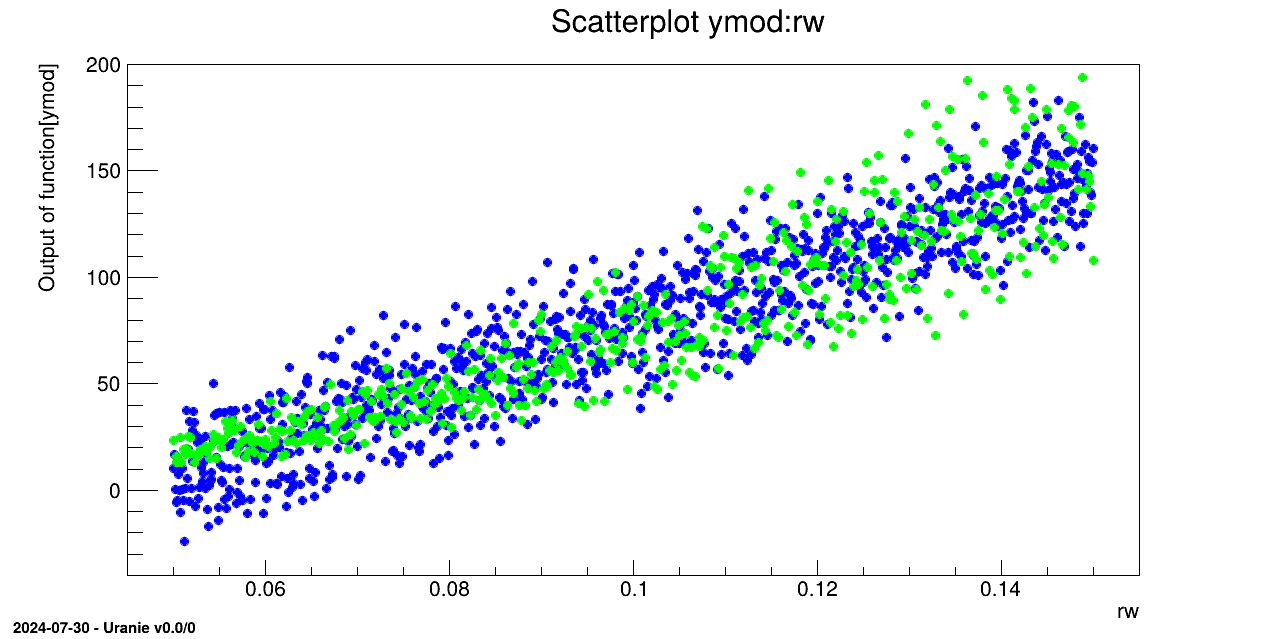
tds2->draw("ymod:rw");
tds->draw("yhat:rw","","same");
}
The TDataServer is loaded with the database contained in the file flowrate_sampler_launcher_500.dat
with the fileDataRead method:
TDataServer * tds = new TDataServer();
tds->fileDataRead("flowrate_sampler_launcher_500.dat");
The linear regression is initialised and characteristic values are computed for each variable with the
estimate method:
TLinearRegression *tlin = new TLinearRegression(tds, "rw:r:tu:tl:hu:hl:l:kw", "yhat");
tlin->estimate();
The model is exported in an external file in C++ language _FileContainingFunction_.C where the function name is
MultiLinearFunction:
tlin->exportFunction("c++", "_FileContainingFunction_", "MultiLinearFunction");
A second TDataServer is created. The previous variables then obey uniform laws and are linked as
TAttribute in this new TDataServer:
tds2->addAttribute( new TUniformDistribution("rw", 0.05, 0.15));
tds2->addAttribute( new TUniformDistribution("r", 100.0, 50000.0));
tds2->addAttribute( new TUniformDistribution("tu", 63070.0, 115600.0));
tds2->addAttribute( new TUniformDistribution("tl", 63.1, 116.0));
tds2->addAttribute( new TUniformDistribution("hu", 990.0, 1110.0));
tds2->addAttribute( new TUniformDistribution("hl", 700.0, 820.0));
tds2->addAttribute( new TUniformDistribution("l", 1120.0, 1680.0));
tds2->addAttribute( new TUniformDistribution("kw", 9855.0, 12045.0));
A sampling is realised with a LHS method on 1000 patterns:
TSampling *sampling = new TSampling(tds2, "lhs", nS=1000);
sampling->generateSample();
The previously exported macro _FileContainingFunction_.C is loaded so as to perform calculation over the database
in the second TDataServer with the function MultiLinearFunction. Results are stored in the ymod variable:
gROOT->LoadMacro("_FileContainingFunction_.C");
TLauncherFunction * tlf = new TLauncherFunction(tds2, "MultiLinearFunction", "", "ymod");
tlf->run();
The objective of this macro is to build a surrogate model (an Artificial Neural Network) from a database. From a
first database created from an ASCII file flowrate_sampler_launcher_500.dat (which defines
values for eight variables described in Section IV.1.2.1 on 500
patterns), two ASCII files are created: one with the 300st patterns
(_flowrate_sampler_launcher_app_.dat), the other with the 200 last patterns
(_flowrate_sampler_launcher_val_.dat). The surrogate model is built with the database
extracted from the first of the two files, the second allowing to perform calculations with a function.
{
// Create a TDataServer
// Load a database in an ASCII file
TDataServer * tdsu = new TDataServer("tdsu","tds u");
tdsu->fileDataRead("flowrate_sampler_launcher_500.dat");
//Create database
tdsu->exportData("_flowrate_sampler_launcher_app_.dat", "","tdsFlowrate__n__iter__<=300");
tdsu->exportData("_flowrate_sampler_launcher_val_.dat", "","tdsFlowrate__n__iter__>300");
TDataServer * tds = new TDataServer("tdsFlowrate", "tds for flowrate");
tds->fileDataRead("_flowrate_sampler_launcher_app_.dat");
TCanvas *c = new TCanvas("c1", "Graph for the Macro modeler",5,64,1270,667);
// Buils a surrogate model (Artificial Neural Networks) from the DataBase
TANNModeler* tann = new TANNModeler(tds, "rw:r:tu:tl:hu:hl:l:kw,3,yhat");
tann->setFcnTol(1e-5);
//tann->setLog();
tann->train(3, 2, "test");
tann->exportFunction("pmmlc++","uranie_ann_flowrate","ANNflowrate");
gROOT->LoadMacro("uranie_ann_flowrate.C");
TDataServer * tdsv = new TDataServer();
tdsv->fileDataRead("_flowrate_sampler_launcher_val_.dat");
cout << tdsv->getNPatterns() << endl;
// evaluate the surrogate model on the database
TLauncherFunction * tlf = new TLauncherFunction(tdsv, "ANNflowrate", "rw:r:tu:tl:hu:hl:l:kw", "yann");
tlf->run();
tdsv->startViewer();
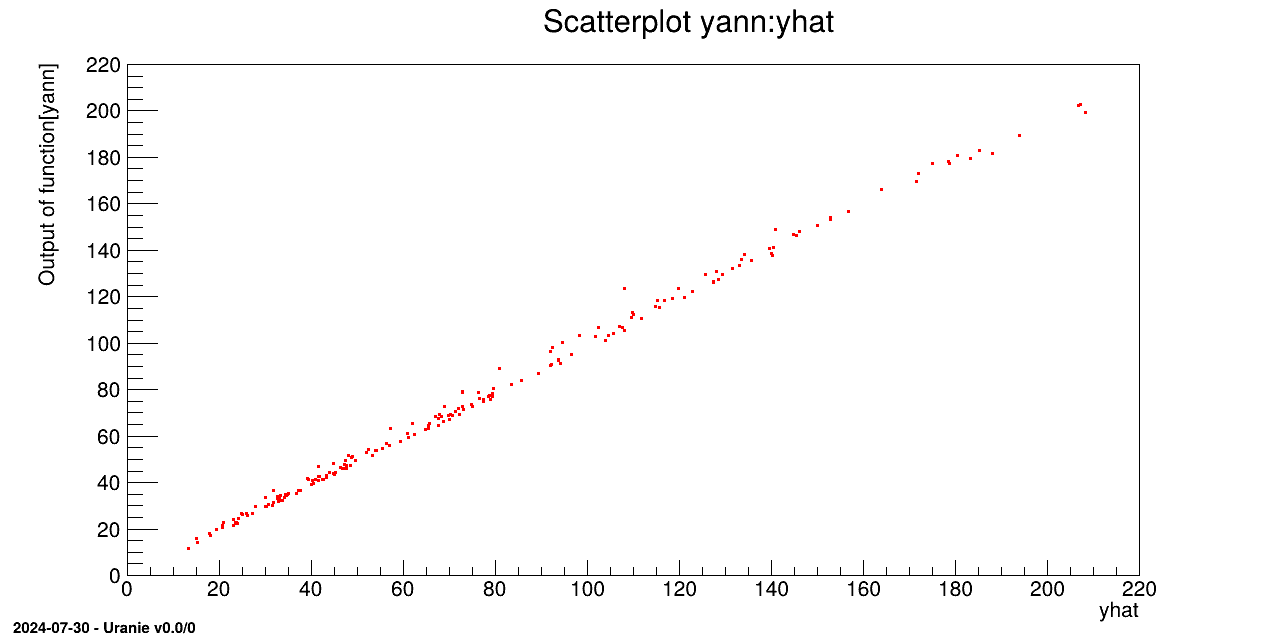
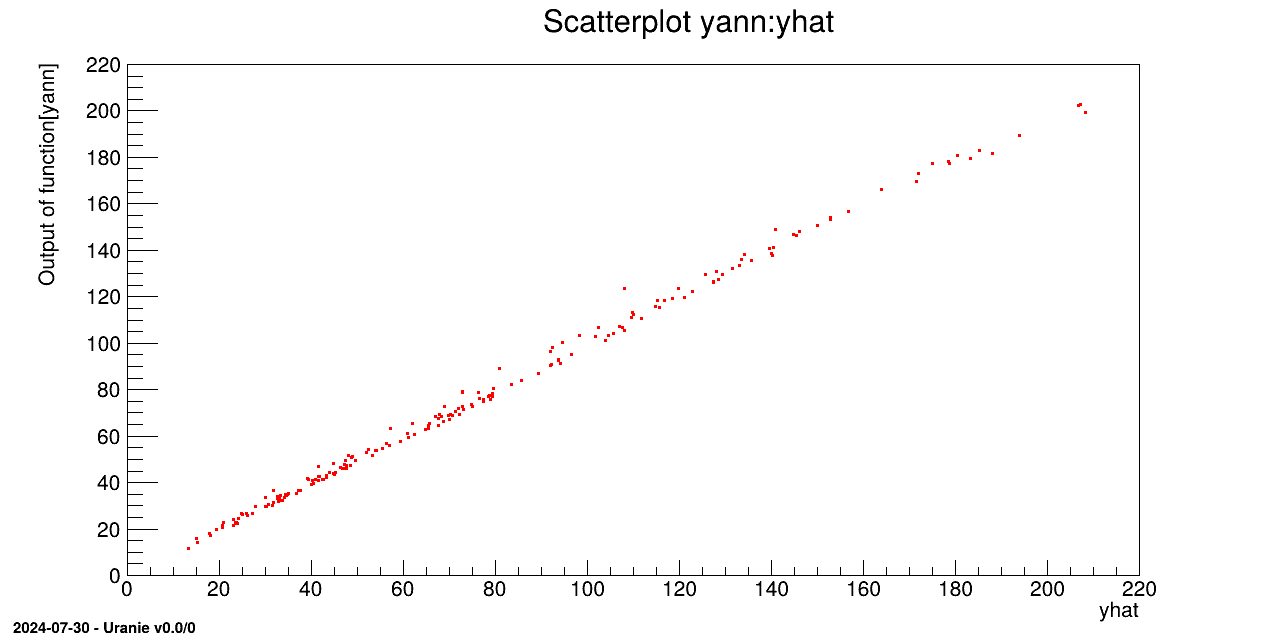
tdsv->Draw("yann:yhat");
// tdsv->draw("yhat");
}
The main TDataServer loads the main ASCII data file flowrate_sampler_launcher_500.dat
TDataServer * tdsu = new TDataServer("tdsu","tds u");
tdsu->fileDataRead("flowrate_sampler_launcher_500.dat");
The database is split in two parts by exporting the 300st patterns in a file and the remaining 200 in another one:
tdsu->exportData("_flowrate_sampler_launcher_app_.dat", "","tdsFlowrate__n__iter__<=300");
tdsu->exportData("_flowrate_sampler_launcher_val_.dat", "","tdsFlowrate__n__iter__>300");
A second TDataServer loads _flowrate_sampler_launcher_app_.dat and builds the surrogate
model over all the variables:
TDataServer * tds = new TDataServer("tds", "tds for flowrate");
tds->fileDataRead("_flowrate_sampler_launcher_app_.dat");
TANNModeler* tann = new TANNModeler(tds, "rw:r:tu:tl:hu:hl:l:kw,3,yhat");
tann->setFcnTol(1e-5);
tann->train(3, 2, "test");
The model is exported in an external file in C++ language "uranie_ann_flowrate.C where the
function name is ANNflowrate:
tann->exportFunction("c++", "uranie_ann_flowrate.C","ANNflowrate");
The model is loaded from the macro "uranie_ann_flowrate.C and applied on the second database
with the function ANNflowrate:
gROOT->LoadMacro("uranie_ann_flowrate.C");
TDataServer * tdsv = new TDataServer();
tdsv->fileDataRead("_flowrate_sampler_launcher_val_.dat");
TLauncherFunction * tlf = new TLauncherFunction(tdsv, "ANNflowrate", "rw:r:tu:tl:hu:hl:l:kw", "yann");
tlf->run();
Processing modelerFlowrateNeuralNetworks.C...
--- Uranie v4.10/0 --- Developed with ROOT (6.32.08)
Copyright (C) 2013-2025 CEA/DES
Contact: support-uranie@cea.fr
Date: Fri Feb 21, 2025
<URANIE::WARNING>
<URANIE::WARNING> *** URANIE WARNING ***
<URANIE::WARNING> *** File[${SOURCEDIR}/dataSERVER/souRCE/TDataServer.cxx] Line[759]
<URANIE::WARNING> TDataServer::fileDataRead: Expected iterator tdsu__n__iter__ not found but tdsFlowrate__n__iter__ looks like an URANIE iterator => Will be used as so.
<URANIE::WARNING> *** END of URANIE WARNING ***
<URANIE::WARNING>
** TANNModeler::train niter[3] ninit[2]
** init the ANN
** Input (1) Name[rw] Min[0.101018] Max[0.0295279]
** Input (2) Name[r] Min[25668.6] Max[14838.3]
** Input (3) Name[tu] Min[89914.4] Max[14909]
** Input (4) Name[tl] Min[89.0477] Max[15.0121]
** Input (5) Name[hu] Min[1048.92] Max[34.8615]
** Input (6) Name[hl] Min[763.058] Max[33.615]
** Input (7) Name[l] Min[1401.09] Max[163.611]
** Input (8) Name[kw] Min[10950.2] Max[635.503]
** Output (1) Name[yhat] Min[78.0931] Max[44.881]
** Tolerance (1e-05)
** sHidden (3) (3)
** Nb Weights (31)
** ActivationFunction[LOGISTIC]
**
** iter[1/3] : *= : mse_min[0.00150425]
** iter[2/3] : ** : mse_min[0.0024702]
** iter[3/3] : *= : mse_min[0.00122167]
** solutions : 3
** isol[1] iter[0] learn[0.00126206] test[0.00150425] *
** isol[2] iter[1] learn[0.00152949] test[0.0024702]
** isol[3] iter[2] learn[0.00107926] test[0.00122167] *
** CPU training finished. Total elapsed time: 1.49 sec
*******************************
*** TModeler::exportFunction lang[pmmlc++] file[uranie_ann_flowrate] name[ANNflowrate] soption[]
*******************************
*******************************
*** exportFunction lang[pmmlc++] file[uranie_ann_flowrate] name[ANNflowrate]
*******************************
PMML Constructor: uranie_ann_flowrate.pmml
*** End Of exportModelPMML
*******************************
*** End Of exportFunction
*******************************
The objective of this macro is to build a surrogate model (an Artificial Neural Network) from a PMML file.
{
// Create a TDataServer
// Load a database in an ASCII file
TDataServer * tds = new TDataServer("tdsFlowrate", "tds for flowrate");
tds->fileDataRead("_flowrate_sampler_launcher_app_.dat");
TCanvas *c = new TCanvas("c1", "Graph for the Macro modeler",5,64,1270,667);
// Build a surrogate model (Artificial Neural Networks) from the PMML file
TANNModeler* tann = new TANNModeler(tds, "uranie_ann_flowrate.pmml","ANNflowrate");
// export the surrogate model in a C file
tann->exportFunction("c++", "uranie_ann_flowrate_loaded","ANNflowrate");
// load the surrogate model in the C file
gROOT->LoadMacro("uranie_ann_flowrate_loaded.C");
TDataServer * tdsv = new TDataServer("tdsv", "tds for surrogate model");
tdsv->fileDataRead("_flowrate_sampler_launcher_val_.dat");
cout << tdsv->getNPatterns() << endl;
// evaluate the surrogate model on the database
TLauncherFunction * tlf = new TLauncherFunction(tdsv, "ANNflowrate", "rw:r:tu:tl:hu:hl:l:kw", "yann");
tlf->run();
tdsv->startViewer();
tdsv->Draw("yann:yhat");
// tdsv->draw("yhat");
}
A TDataServer loads _flowrate_sampler_launcher_app_.dat:
TDataServer * tds = new TDataServer("tds", "tds for flowrate");
tds->fileDataRead("_flowrate_sampler_launcher_app_.dat");
The surrogate model is loaded from a PMML file:
TANNModeler* tann = new TANNModeler(tds, "uranie_ann_flowrate.pmml","ANNflowrate");
The model is exported in an external file in C++ language "uranie_ann_flowrate.C where the
function name is ANNflowrate:
tann->exportFunction("c++", "uranie_ann_flowrate.C","ANNflowrate");
The model is loaded from the macro "uranie_ann_flowrate.C and applied on the database with
the function ANNflowrate:
gROOT->LoadMacro("uranie_ann_flowrate.C");
TDataServer * tdsv = new TDataServer();
tdsv->fileDataRead("_flowrate_sampler_launcher_val_.dat");
TLauncherFunction * tlf = new TLauncherFunction(tdsv, "ANNflowrate", "rw:r:tu:tl:hu:hl:l:kw", "yann");
tlf->run();
Processing modelerFlowrateNeuralNetworksLoadingPMML.C...
--- Uranie v4.10/0 --- Developed with ROOT (6.32.08)
Copyright (C) 2013-2025 CEA/DES
Contact: support-uranie@cea.fr
Date: Fri Feb 21, 2025
PMML Constructor: uranie_ann_flowrate.pmml
*******************************
*** TModeler::exportFunction lang[c++] file[uranie_ann_flowrate_loaded] name[ANNflowrate] soption[]
*******************************
*******************************
*** exportFunction lang[c++] file[uranie_ann_flowrate_loaded] name[ANNflowrate]
*** End Of exportFunction
*******************************
The objective of this macro is to build a surrogate model (an Artificial Neural Network) from a database for a
"Classification" Problem. From a first database loaded from the ASCII file
problem2Classes_001000.dat, which defines three variables 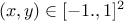 and
and
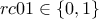 on 1000 patterns,
on 1000 patterns,
 |
two ASCII files are created, one with the 800st patterns (_problem2Classes_app_.dat), the other with the 200 last patterns
(_problem2Classes_val_.dat). The surrogate model is built with the database
extracted from the first of the two files, the second allowing to perform calculations with a function.
{
Int_t nH1 = 15;
Int_t nH2 = 10;
Int_t nH3 = 5;
TString sX = "x:y";
TString sY = "rc01";
TString sYhat = Form("%shat_%d_%d_%d", sY.Data(), nH1, nH2, nH3);
// Load a database in an ASCII file
TDataServer * tdsu = new TDataServer("tdsu","tds u");
tdsu->fileDataRead("problem2Classes_001000.dat");
// Split into 2 datasets for learning and testing
tdsu->exportData("_problem2Classes_app_.dat", "", Form("%s<=800", tdsu->getIteratorName()));
tdsu->exportData("_problem2Classes_val_.dat", "", Form("%s>800", tdsu->getIteratorName()));
TDataServer * tds = new TDataServer("tdsApp", "tds App for problem2Classes");
tds->fileDataRead("_problem2Classes_app_.dat");
TANNModeler* tann = new TANNModeler(tds, Form("%s,%d,%d,%d,@%s", sX.Data(), nH1, nH2, nH3, sY.Data()));
//tann->setLog();
tann->setNormalization(TANNModeler::kMinusOneOne);
tann->setFcnTol(1e-6);
tann->train(2, 2, "test", kFALSE);
tann->exportFunction("c++","uranie_ann_problem2Classes","ANNproblem2Classes");
gROOT->LoadMacro("uranie_ann_problem2Classes.C");
TDataServer * tdsv = new TDataServer();
tdsv->fileDataRead("_problem2Classes_val_.dat");
// evaluate the surrogate model on the database
TLauncherFunction * tlf = new TLauncherFunction(tdsv, "ANNproblem2Classes", sX, sYhat);
tlf->run();
TCanvas *c = new TCanvas("c1", "Graph for the Macro modeler",5,64,1270,667);
// Buils a surrogate model (Artificial Neural Networks) from the DataBase
tds->getTuple()->SetMarkerStyle(8);
tds->getTuple()->SetMarkerSize(1.0);
tds->getTuple()->SetMarkerColor(kGreen);
tds->draw(Form("%s:%s", sY.Data(), sX.Data()));
tdsv->getTuple()->SetMarkerStyle(8);
tdsv->getTuple()->SetMarkerSize(0.75);
tdsv->getTuple()->SetMarkerColor(kRed);
tdsv->Draw(Form("%s:%s", sYhat.Data(), sX.Data()),"","same");
}
The main TDataServer loads the main ASCII data file problem2Classes_001000.dat
TDataServer * tdsu = new TDataServer("tdsu","tds u");
tdsu->fileDataRead("problem2Classes_001000.dat");
The database is split with the internal iterator attribute in two parts by exporting the 800st patterns in a file and the remaining 200 in another one
tdsu->exportData("_problem2Classes_app_.dat", "",Form("%s<=800", tdsu->getIteratorName()));
tdsu->exportData("_problem2Classes_val_.dat", "", Form("%s>800", tdsu->getIteratorName()));
A second TDataServer loads _problem2Classes_app_.dat and builds the surrogate model over all the variables
with 3 hidden layers, a Hyperbolic Tangent (TanH) activation function (normalization) in the 3 hidden layers and set the function tolerance to 1e-.6. The "@" character behind the output name defines a classification problem.
TDataServer * tds = new TDataServer("tds", "tds for the 2 Classes problem");
tds->fileDataRead("_problem2Classes_app_.dat");
TANNModeler* tann = new TANNModeler(tds, Form("%s,%d,%d,%d,@%s", sX.Data(), nH1, nH2, nH3, sY.Data()));
tann->setNormalization(TANNModeler::kMinusOneOne);
tann->setFcnTol(1e-6);
tann->train(3, 2, "test");
The model is exported in an external file in C++ language "uranie_ann_problem2Classes.C" where the
function name is ANNproblem2Classes
tann->exportFunction("c++", "uranie_ann_problem2Classes.C","ANNproblem2Classes");
The model is loaded from the macro "uranie_ann_problem2Classes.C and applied on the second database
with the function ANNproblem2Classes.
gROOT->LoadMacro("uranie_ann_problem2Classes.C");
TDataServer * tdsv = new TDataServer();
tdsv->fileDataRead("_problem2Classes_val_.dat");
TLauncherFunction * tlf = new TLauncherFunction(tdsv, "ANNproblem2Classes", sX, sYhat);
tlf->run();
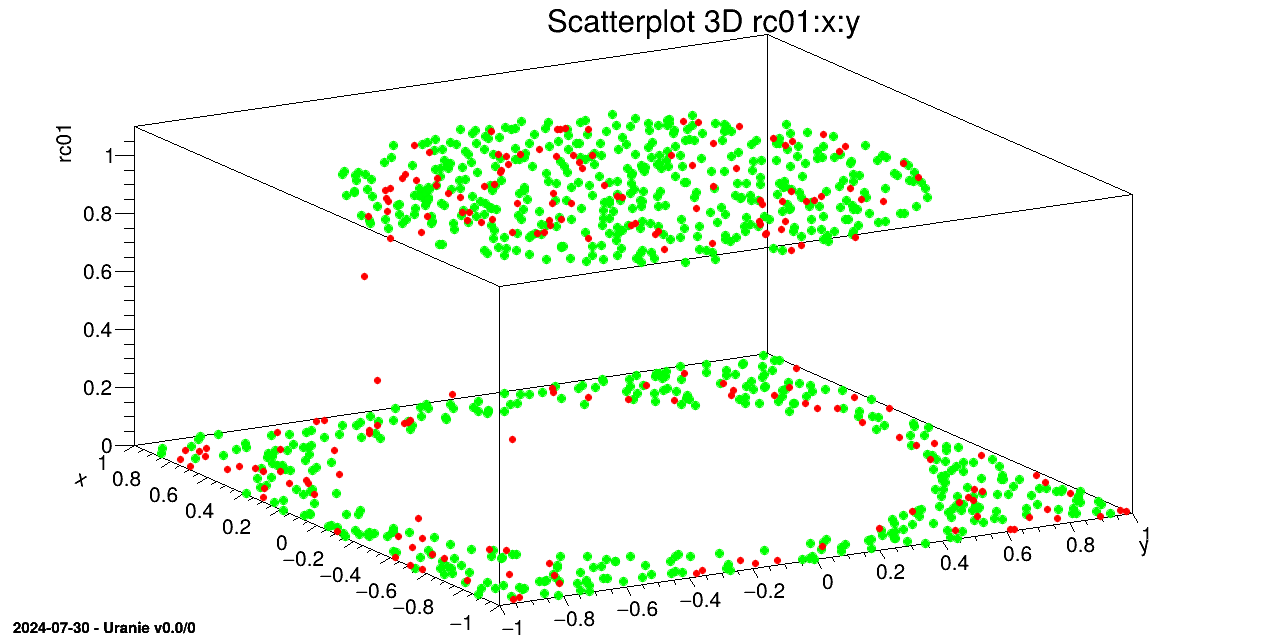
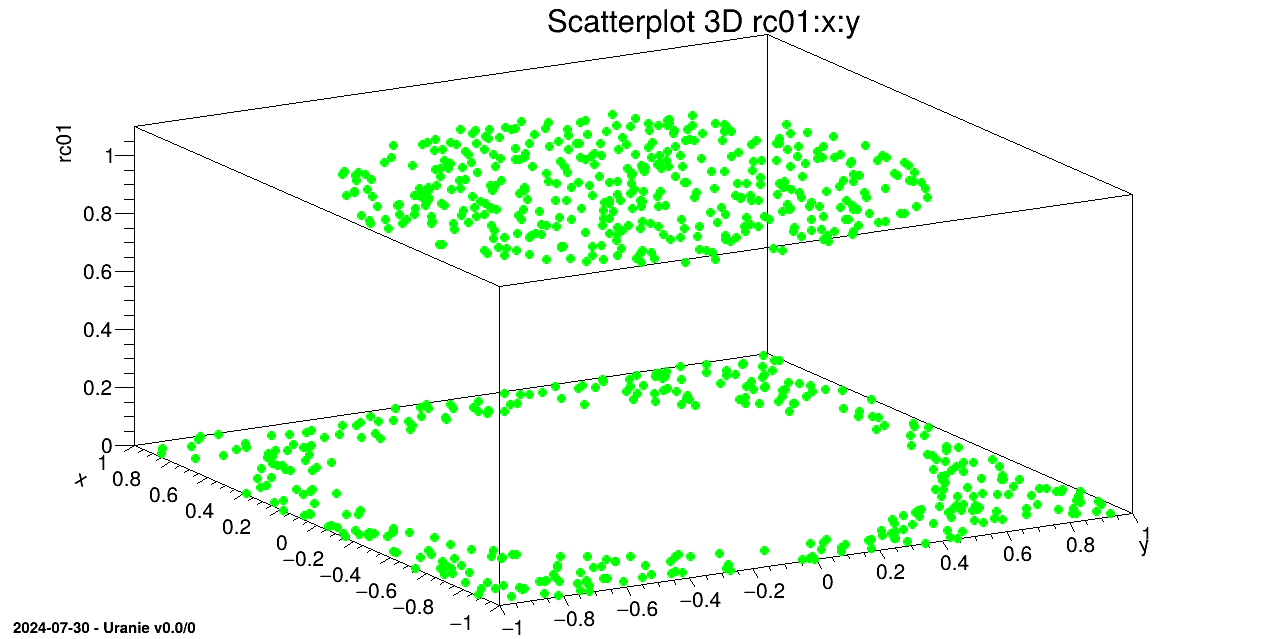
We draw on a 3D graph, the learning database (in green) and the estimations by the Artificial Neural Network with red points.
TCanvas *c = new TCanvas("c1", "Graph for the Macro modeler",5,64,1270,667);
tds->getTuple()->SetMarkerStyle(8);
tds->getTuple()->SetMarkerSize(1.0);
tds->getTuple()->SetMarkerColor(kGreen);
tds->draw(Form("%s:%s", sY.Data(), sX.Data()));
tdsv->getTuple()->SetMarkerStyle(8);
tdsv->getTuple()->SetMarkerSize(0.75);
tdsv->getTuple()->SetMarkerColor(kRed);
tdsv->Draw(Form("%s:%s", sYhat.Data(), sX.Data()),"","same");
Processing modelerClassificationNeuralNetworks.C...
--- Uranie v4.10/0 --- Developed with ROOT (6.32.08)
Copyright (C) 2013-2025 CEA/DES
Contact: support-uranie@cea.fr
Date: Fri Feb 21, 2025
<URANIE::WARNING>
<URANIE::WARNING> *** URANIE WARNING ***
<URANIE::WARNING> *** File[${SOURCEDIR}/dataSERVER/souRCE/TDataServer.cxx] Line[759]
<URANIE::WARNING> TDataServer::fileDataRead: Expected iterator tdsu__n__iter__ not found but _tds___n__iter__ looks like an URANIE iterator => Will be used as so.
<URANIE::WARNING> *** END of URANIE WARNING ***
<URANIE::WARNING>
<URANIE::WARNING>
<URANIE::WARNING> *** URANIE WARNING ***
<URANIE::WARNING> *** File[${SOURCEDIR}/dataSERVER/souRCE/TDataServer.cxx] Line[759]
<URANIE::WARNING> TDataServer::fileDataRead: Expected iterator tdsApp__n__iter__ not found but _tds___n__iter__ looks like an URANIE iterator => Will be used as so.
<URANIE::WARNING> *** END of URANIE WARNING ***
<URANIE::WARNING>
** TANNModeler::train niter[2] ninit[2]
** init the ANN
** Input (1) Name[x] Min[-0.999842] Max[0.998315]
** Input (2) Name[y] Min[-0.999434] Max[0.998071]
** Output (1) Name[rc01] Min[0] Max[1]
** Tolerance (1e-06)
** sHidden (15,10,5) (30)
** Nb Weights (266)
** ActivationFunction[TanH]
**
** iter[1/2] : ** : mse_min[0.00813999]
** iter[2/2] : ** : mse_min[0.00636923]
** solutions : 2
** isol[1] iter[0] learn[0.00405863] test[0.00813999] *
** isol[2] iter[1] learn[0.00560685] test[0.00636923] *
** CPU training finished. Total elapsed time: 17 sec
*******************************
*** TModeler::exportFunction lang[c++] file[uranie_ann_problem2Classes] name[ANNproblem2Classes] soption[]
*******************************
*******************************
*** exportFunction lang[c++] file[uranie_ann_problem2Classes] name[ANNproblem2Classes]
*** End Of exportFunction
*******************************
The objective of this macro is to build a polynomial chaos expansion in order to get a surrogate model along with a
global sensitivity interpretation for the flowrate function, whose purpose and behaviour have been
already introduced in Section IV.1.2.1. The method used here is
the regression one, as discussed below.
{
gROOT->LoadMacro("UserFunctions.C");
// Define the DataServer
TDataServer *tds = new TDataServer("tdsFlowrate", "Design of experiments for Flowrate");
// Add the eight attributes of the study with uniform law
tds->addAttribute( new TUniformDistribution("rw", 0.05, 0.15));
tds->addAttribute( new TUniformDistribution("r", 100.0, 50000.0));
tds->addAttribute( new TUniformDistribution("tu", 63070.0, 115600.0));
tds->addAttribute( new TUniformDistribution("tl", 63.1, 116.0));
tds->addAttribute( new TUniformDistribution("hu", 990.0, 1110.0));
tds->addAttribute( new TUniformDistribution("hl", 700.0, 820.0));
tds->addAttribute( new TUniformDistribution("l", 1120.0, 1680.0));
tds->addAttribute( new TUniformDistribution("kw", 9855.0, 12045.0));
// Define of TNisp object
TNisp *nisp = new TNisp(tds);
nisp->generateSample("QmcSobol", 500); //State that there is a sample ...
// Compute the output variable
TLauncherFunction * tlf = new TLauncherFunction(tds, "flowrateModel","*","ymod");
tlf->setDrawProgressBar(kFALSE);
tlf->run();
// build a chaos polynomial
TPolynomialChaos *pc = new TPolynomialChaos(tds,nisp);
// compute the pc coefficients using the "Regression" method
Int_t degree = 4;
pc->setDegree(degree);
pc->computeChaosExpansion("Regression");
// Uncertainty and sensitivity analysis
cout << "Variable ymod ================" << endl;
cout << "Mean = " << pc->getMean("ymod") << endl;
cout << "Variance = " << pc->getVariance("ymod") << endl;
cout << "First Order Indices ================" << endl;
cout << "Indice First Order[1] = " << pc->getIndexFirstOrder(0,0) << endl;
cout << "Indice First Order[2] = " << pc->getIndexFirstOrder(1,0) << endl;
cout << "Indice First Order[3] = " << pc->getIndexFirstOrder(2,0) << endl;
cout << "Indice First Order[4] = " << pc->getIndexFirstOrder(3,0) << endl;
cout << "Indice First Order[5] = " << pc->getIndexFirstOrder(4,0) << endl;
cout << "Indice First Order[6] = " << pc->getIndexFirstOrder(5,0) << endl;
cout << "Indice First Order[7] = " << pc->getIndexFirstOrder(6,0) << endl;
cout << "Indice First Order[8] = " << pc->getIndexFirstOrder(7,0) << endl;
cout << "Total Order Indices ================" << endl;
cout << "Indice Total Order[1] = " << pc->getIndexTotalOrder("rw","ymod") << endl;
cout << "Indice Total Order[2] = " << pc->getIndexTotalOrder("r","ymod") << endl;
cout << "Indice Total Order[3] = " << pc->getIndexTotalOrder("tu","ymod") << endl;
cout << "Indice Total Order[4] = " << pc->getIndexTotalOrder("tl","ymod") << endl;
cout << "Indice Total Order[5] = " << pc->getIndexTotalOrder("hu","ymod") << endl;
cout << "Indice Total Order[6] = " << pc->getIndexTotalOrder("hl","ymod") << endl;
cout << "Indice Total Order[7] = " << pc->getIndexTotalOrder("l","ymod") << endl;
cout << "Indice Total Order[8] = " << pc->getIndexTotalOrder("kw","ymod") << endl;
// Dump main factors up to a certain threshold
Double_t seuil = 0.99;
cout<<"Ordered functionnal ANOVA"<<endl;
pc->getAnovaOrdered(seuil,0);
cout << "Number of experiments = " << tds->getNPatterns() << endl;
//save the pv in a program (C langage)
pc->exportFunction("NispFlowrate","NispFlowrate");
}
The first part is just creating a TDataServer and providing the attributes needed to define the problem. From there, a
TNisp object is created, providing the dataserver that specifies the inputs. This class is
used to generate the sample.
// Define of TNisp object
TNisp *nisp = new TNisp(tds);
nisp->generateSample("QmcSobol", 500); //State that there is a sample ...
The function is launched through a TLauncherFunction instance in order to get the output
values that will be needed to train the surrogate model.
TLauncherFunction * tlf = new TLauncherFunction(tds, "flowrateModel","*","ymod");
tlf->run();
Finally, a TPolynomialChaos instance is created and the computation of the coefficients is
performed by requesting a truncature on the resulting degree of the polynomial expansion (set to 4) and the use of
a regression method.
// build a chaos polynomial
TPolynomialChaos *pc = new TPolynomialChaos(tds,nisp);
// compute the pc coefficients using the "Regression" method
Int_t degree = 4;
pc->setDegree(degree);
pc->computeChaosExpansion("Regression");
The rest of the code is showing how to access the resulting sensitivity indices either one-by-one, or ordered up to a chosen threshold of the output variance.
Processing modelerFlowratePolynChaosRegression.C...
--- Uranie v4.10/0 --- Developed with ROOT (6.32.08)
Copyright (C) 2013-2025 CEA/DES
Contact: support-uranie@cea.fr
Date: Fri Feb 21, 2025
Variable ymod ================
Mean = 77.651
Variance = 2079.68
First Order Indices ================
Indice First Order[1] = 0.828732
Indice First Order[2] = 1.05548e-06
Indice First Order[3] = 9.86805e-08
Indice First Order[4] = 4.77668e-06
Indice First Order[5] = 0.0414015
Indice First Order[6] = 0.0413215
Indice First Order[7] = 0.039401
Indice First Order[8] = 0.00954644
Total Order Indices ================
Indice Total Order[1] = 0.866773
Indice Total Order[2] = 9.27467e-06
Indice Total Order[3] = 7.01768e-06
Indice Total Order[4] = 1.71944e-05
Indice Total Order[5] = 0.0541916
Indice Total Order[6] = 0.0540578
Indice Total Order[7] = 0.0522121
Indice Total Order[8] = 0.0127809
The objective of this macro is to build a polynomial chaos expansion in order to get a surrogate model along with a
global sensitivity interpretation for the flowrate function, whose purpose and behaviour have been
already introduced in Section IV.1.2.1. The method used here is
the regression one, as discussed below.
{
gROOT->LoadMacro("UserFunctions.C");
// Define the DataServer
TDataServer *tds = new TDataServer("tdsFlowrate", "Design of experiments for Flowrate");
// Add the eight attributes of the study with uniform law
tds->addAttribute( new TUniformDistribution("rw", 0.05, 0.15));
tds->addAttribute( new TUniformDistribution("r", 100.0, 50000.0));
tds->addAttribute( new TUniformDistribution("tu", 63070.0, 115600.0));
tds->addAttribute( new TUniformDistribution("tl", 63.1, 116.0));
tds->addAttribute( new TUniformDistribution("hu", 990.0, 1110.0));
tds->addAttribute( new TUniformDistribution("hl", 700.0, 820.0));
tds->addAttribute( new TUniformDistribution("l", 1120.0, 1680.0));
tds->addAttribute( new TUniformDistribution("kw", 9855.0, 12045.0));
// Define of TNisp object
TNisp *nisp = new TNisp(tds);
nisp->generateSample("Petras", 5); //State that there is a sample ...
// Compute the output variable
TLauncherFunction * tlf = new TLauncherFunction(tds, "flowrateModel","*","ymod");
tlf->setDrawProgressBar(kFALSE);
tlf->run();
// build a chaos polynomial
TPolynomialChaos *pc = new TPolynomialChaos(tds,nisp);
// compute the pc coefficients using the "Integration" method
Int_t degree = 4;
pc->setDegree(degree);
pc->computeChaosExpansion("Integration");
// Uncertainty and sensitivity analysis
cout << "Variable ymod ================" << endl;
cout << "Mean = " << pc->getMean("ymod") << endl;
cout << "Variance = " << pc->getVariance("ymod") << endl;
cout << "First Order Indices ================" << endl;
cout << "Indice First Order[1] = " << pc->getIndexFirstOrder(0,0) << endl;
cout << "Indice First Order[2] = " << pc->getIndexFirstOrder(1,0) << endl;
cout << "Indice First Order[3] = " << pc->getIndexFirstOrder(2,0) << endl;
cout << "Indice First Order[4] = " << pc->getIndexFirstOrder(3,0) << endl;
cout << "Indice First Order[5] = " << pc->getIndexFirstOrder(4,0) << endl;
cout << "Indice First Order[6] = " << pc->getIndexFirstOrder(5,0) << endl;
cout << "Indice First Order[7] = " << pc->getIndexFirstOrder(6,0) << endl;
cout << "Indice First Order[8] = " << pc->getIndexFirstOrder(7,0) << endl;
cout << "Total Order Indices ================" << endl;
cout << "Indice Total Order[1] = " << pc->getIndexTotalOrder("rw","ymod") << endl;
cout << "Indice Total Order[2] = " << pc->getIndexTotalOrder("r","ymod") << endl;
cout << "Indice Total Order[3] = " << pc->getIndexTotalOrder("tu","ymod") << endl;
cout << "Indice Total Order[4] = " << pc->getIndexTotalOrder("tl","ymod") << endl;
cout << "Indice Total Order[5] = " << pc->getIndexTotalOrder("hu","ymod") << endl;
cout << "Indice Total Order[6] = " << pc->getIndexTotalOrder("hl","ymod") << endl;
cout << "Indice Total Order[7] = " << pc->getIndexTotalOrder("l","ymod") << endl;
cout << "Indice Total Order[8] = " << pc->getIndexTotalOrder("kw","ymod") << endl;
// Dump main factors up to a certain threshold
Double_t seuil = 0.99;
cout<<"Ordered functionnal ANOVA"<<endl;
pc->getAnovaOrdered(seuil,0);
cout << "Number of experiments = " << tds->getNPatterns() << endl;
//save the pv in a program (C langage)
pc->exportFunction("NispFlowrate","NispFlowrate");
}
The first part is just creating a TDataServer and providing the attributes needed to define the problem. From there, a
TNisp object is created, providing the dataserver that specifies the inputs. This class is
used to generate the sample (Petras being a design-of-experiments dedicated to integration problem).
// Define of TNisp object
TNisp *nisp = new TNisp(tds);
nisp->generateSample("Petras", 5); //State that there is a sample ...
The function is launched through a TLauncherFunction instance in order to get the output
values that will be needed to train the surrogate model.
TLauncherFunction * tlf = new TLauncherFunction(tds, "flowrateModel","*","ymod");
tlf->run();
Finally, a TPolynomialChaos instance is created and the computation of the coefficients is
performed by requesting a truncature on the resulting degree of the polynomial expansion (set to 4) and the use of
a regression method.
// build a chaos polynomial
TPolynomialChaos *pc = new TPolynomialChaos(tds,nisp);
// compute the pc coefficients using the "Integration" method
Int_t degree = 4;
pc->setDegree(degree);
pc->computeChaosExpansion("Integration");
The rest of the code is showing how to access the resulting sensitivity indices either one-by-one, or ordered up to a chosen threshold of the output variance.
Processing modelerFlowratePolynChaosIntegration.C...
--- Uranie v4.10/0 --- Developed with ROOT (6.32.08)
Copyright (C) 2013-2025 CEA/DES
Contact: support-uranie@cea.fr
Date: Fri Feb 21, 2025
Variable ymod ================
Mean = 77.6511
Variance = 2078.91
First Order Indices ================
Indice First Order[1] = 0.828922
Indice First Order[2] = 1.04477e-06
Indice First Order[3] = 8.94692e-12
Indice First Order[4] = 5.30299e-06
Indice First Order[5] = 0.0413852
Indice First Order[6] = 0.0413852
Indice First Order[7] = 0.0393419
Indice First Order[8] = 0.00952157
Total Order Indices ================
Indice Total Order[1] = 0.866834
Indice Total Order[2] = 2.16178e-06
Indice Total Order[3] = 2.28296e-09
Indice Total Order[4] = 1.07386e-05
Indice Total Order[5] = 0.0541095
Indice Total Order[6] = 0.0541095
Indice Total Order[7] = 0.0520767
Indice Total Order[8] = 0.0127306
This macro is the one described in Section V.6.2.2, that creates a
simple gaussian process, whose training (utf_4D_train.dat) and testing
(utf_4D_test.dat) database can both be found in the document folder of the Uranie
installation (${URANIESYS}/share/uranie/docUMENTS).
{
// Load observations
TDataServer *tdsObs = new TDataServer("tdsObs", "observations");
tdsObs->fileDataRead("utf_4D_train.dat");
// Construct the GPBuilder
TGPBuilder *gpb = new TGPBuilder(tdsObs, // observations data
"x1:x2:x3:x4", // list of input variables
"y", // output variable
"matern3/2"); // name of the correlation function
// Search for the optimal hyper-parameters
gpb->findOptimalParameters("ML", // optimisation criterion
100, // screening design size
"neldermead", // optimisation algorithm
500); // max. number of optimisation iterations
// Construct the kriging model
TKriging *krig = gpb->buildGP();
// Display model information
krig->printLog();
}This is the result of the last command:
*******************************
** TKriging::printLog[]
*******************************
Input Variables: x1:x2:x3:x4
Output Variable: y
Deterministic trend:
Correlation function: URANIE::Modeler::TMatern32CorrFunction
Correlation length: normalised (not normalised)
1.6181e+00 (1.6172e+00 )
1.4372e+00 (1.4370e+00 )
1.5026e+00 (1.5009e+00 )
6.7884e+00 (6.7944e+00 )
Variance of the gaussian process: 70.8755
RMSE (by Leave One Out): 0.499108
Q2: 0.849843
*******************************
This macro is the one described in Section V.6.2.7, that creates
a gaussian process, whose training (utf_4D_train.dat) and testing
(utf_4D_test.dat) database can both be found in the document folder of the Uranie
installation (${URANIESYS}/share/uranie/docUMENTS), and whose starting point, in the parameter
space, is specified.
{
// Load observations
TDataServer *tdsObs = new TDataServer("tdsObs", "observations");
tdsObs->fileDataRead("utf_4D_train.dat");
// Construct the GPBuilder
TGPBuilder *gpb = new TGPBuilder(tdsObs, "x1:x2:x3:x4", "y", "matern3/2");
// Set the correlation function parameters
Double_t params[4] = {1.0, 0.25, 0.01, 0.3};
gpb->getCorrFunction()->setParameters(params);
// Find the optimal parameters
gpb->findOptimalParameters("ML", // optimisation criterion
0, // screening size MUST be equal to 0
"neldermead", // optimisation algorithm
500, // max. number of optimisation iterations
kFALSE); // we don't reset the parameters of the GP builder
// Create the kriging model
TKriging *krig = gpb->buildGP();
// Display model information
krig->printLog();
}This is the result of the last command:
*******************************
** TKriging::printLog[]
*******************************
Input Variables: x1:x2:x3:x4
Output Variable: y
Deterministic trend:
Correlation function: URANIE::Modeler::TMatern32CorrFunction
Correlation length: normalised (not normalised)
1.6182e+00 (1.6173e+00 )
1.4373e+00 (1.4371e+00 )
1.5027e+00 (1.5011e+00 )
6.7895e+00 (6.7955e+00 )
Variance of the gaussian process: 70.8914
RMSE (by Leave One Out): 0.49911
Q2: 0.849842
*******************************
This macro is the one described in Section V.6.2.4, that creates a gaussian process with a specific trend and an a priori knowledge on the mean and variance of the trend parameters.
{
// Load observations
TDataServer *tdsObs = new TDataServer("tdsObs", "observations");
tdsObs->fileDataRead("utf_4D_train.dat");
// Construct the GPBuilder
TGPBuilder *gpb = new TGPBuilder(tdsObs, // observations data
"x1:x2:x3:x4", // list of input variables
"y", // output variable
"matern3/2", // name of the correlation function
"linear"); // trend defined by a keyword
// Bayesian study
Double_t meanPrior[5] = {0.0, 0.0, -1.0, 0.0, -0.1};
Double_t covPrior[25] = {1e-4, 0.0 , 0.0 , 0.0 , 0.0 ,
0.0 , 1e-4, 0.0 , 0.0 , 0.0 ,
0.0 , 0.0 , 1e-4, 0.0 , 0.0 ,
0.0 , 0.0 , 0.0 , 1e-4, 0.0 ,
0.0 , 0.0 , 0.0 , 0.0 , 1e-4};
gpb->setPriorData(meanPrior, covPrior);
// Search for the optimal hyper-parameters
gpb->findOptimalParameters("ReML", // optimisation criterion
100, // screening design size
"neldermead", // optimisation algorithm
500); // max. number of optimisation iterations
// Construct the kriging model
TKriging *krig = gpb->buildGP();
// Display model information
krig->printLog();
}This is the result of the last command:
*******************************
** TKriging::printLog[]
*******************************
Input Variables: x1:x2:x3:x4
Output Variable: y
Deterministic trend: linear
Trend parameters (5): [3.06586494e-05; 1.64887174e-05; -9.99986787e-01; 1.51959859e-05; -9.99877606e-02 ]
Correlation function: URANIE::Modeler::TMatern32CorrFunction
Correlation length: normalised (not normalised)
2.1450e+00 (2.1438e+00 )
1.9092e+00 (1.9090e+00 )
2.0062e+00 (2.0040e+00 )
8.4315e+00 (8.4390e+00 )
Variance of the gaussian process: 155.533
RMSE (by Leave One Out): 0.495448
Q2: 0.852037
*******************************
This macro is the one described in Section V.6.3.1, to create
and use a simple gaussian process, whose training (utf_4D_train.dat) and testing
(utf_4D_test.dat) database can both be found in the document folder of the Uranie
installation (${URANIESYS}/share/uranie/docUMENTS). It uses the simple one-by-one approch described in
the [metho] for completness.
{
// Load observations
TDataServer *tdsObs = new TDataServer("tdsObs", "observations");
tdsObs->fileDataRead("utf_4D_train.dat");
// Construct the GPBuilder
TGPBuilder *gpb = new TGPBuilder(tdsObs, // observations data
"x1:x2:x3:x4", // list of input variables
"y", // output variable
"matern3/2"); // name of the correlation function
// Search for the optimal hyper-parameters
gpb->findOptimalParameters("ML", // optimisation criterion
100, // screening design size
"neldermead", // optimisation algorithm
500); // max. number of optimisation iterations
// Construct the kriging model
TKriging *krig = gpb->buildGP();
// Display model information
krig->printLog();
// Load the data to estimate
TDataServer *tdsEstim = new TDataServer("tdsEstim", "estimations");
tdsEstim->fileDataRead("utf_4D_test.dat");
// Construction of the launcher
TLauncher2 *lanceur = new TLauncher2(tdsEstim, // data to estimate
krig, // model used for the estimation
"x1:x2:x3:x4", // list of the input variables
"yEstim:vEstim"); // name given to the model's outputs
// Launch the estimations
lanceur->solverLoop();
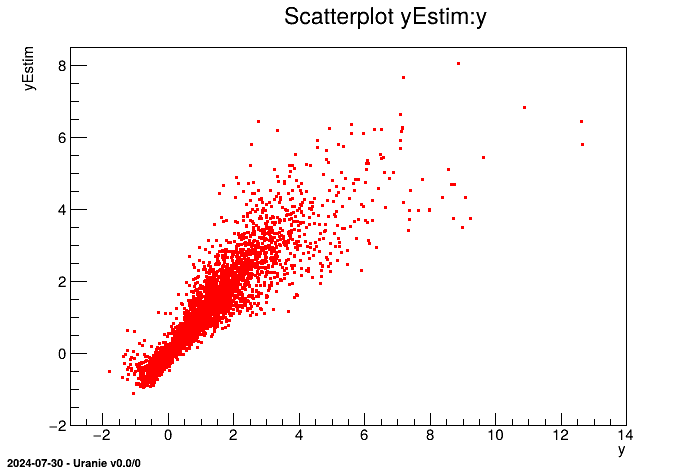
// Display some results
tdsEstim->draw("yEstim:y");
}
This macro is the one described in Section V.6.3.2, to create
and use a simple gaussian process, whose training (utf_4D_train.dat) and testing
(utf_4D_test.dat) database can both be found in the document folder of the Uranie
installation (${URANIESYS}/share/uranie/docUMENTS). It uses the global approach, computing the input
covariance matrix and translating it to the prediction covariance matrix.
{
// Load observations
TDataServer *tdsObs = new TDataServer("tdsObs", "observations");
tdsObs->fileDataRead("utf_4D_train.dat");
// Construct the GPBuilder
TGPBuilder *gpb = new TGPBuilder(tdsObs, // observations data
"x1:x2:x3:x4", // list of input variables
"y", // output variable
"matern1/2"); // name of the correlation function
// Search for the optimal hyper-parameters
gpb->findOptimalParameters("ML", // optimisation criterion
100, // screening design size
"neldermead", // optimisation algorithm
500); // max. number of optimisation iterations
// Construct the kriging model
TKriging *krig = gpb->buildGP();
// Display model information
krig->printLog();
// Load the data to estimate
TDataServer *tdsEstim = new TDataServer("tdsEstim", "estimations");
tdsEstim->fileDataRead("utf_4D_test.dat");
// Reducing the database to 1000 first event (prediction cov matrix of a million value !)
int nST=1000;
tdsEstim->exportData("utf_4D_test_red.dat","",Form("tdsEstim__n__iter__<=%d",nST));
tdsEstim->fileDataRead("utf_4D_test_red.dat",false,true); // Reload reduce sample
krig->estimateWithCov(tdsEstim, // data to estimate
"x1:x2:x3:x4",// list of the input variables
"yEstim:vEstim", // name given to the model's outputs
"y", //name of the true reference if validation database
""); //options
TCanvas *c2=NULL;
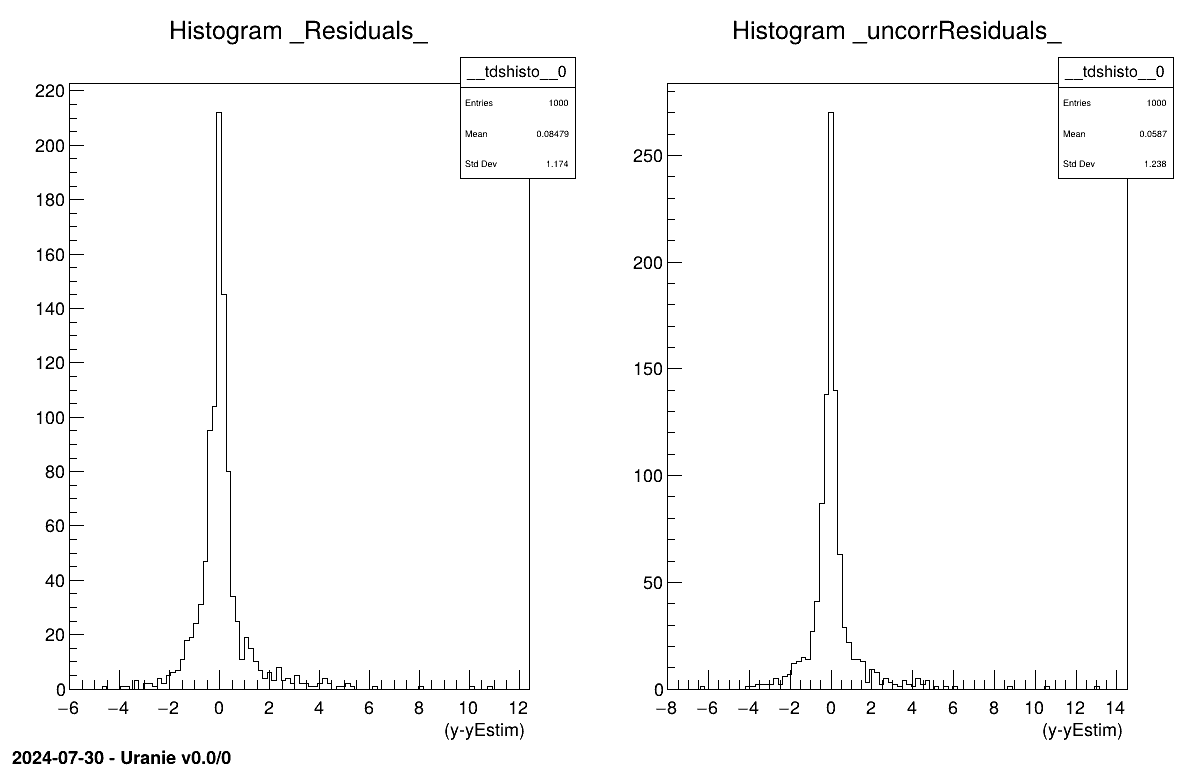
// Residuals plots if true information provided
if( tdsEstim->isAttribute("_Residuals_") )
{
c2 = new TCanvas("c2","c2",1200,800);
c2->Divide(2,1);
c2->cd(1);
// Usual residual considering uncorrated input points
tdsEstim->Draw("_Residuals_");
c2->cd(2);
// Corrected residuals, with prediction covariance matrix
tdsEstim->Draw("_uncorrResiduals_");
}
// Retrieve all the prediction covariance coefficient
tdsEstim->getTuple()->SetEstimate( nST * nST ); //allocate the correct size
// Get a pointer to all values
tdsEstim->getTuple()->Draw("_CovarianceMatrix_","","goff");
double *cov=tdsEstim->getTuple()->GetV1();
//Put these in a matrix nicely created
TMatrixD Cov(nST,nST);
Cov.Use(0,nST-1,0,nST-1,cov);
//Print it if size is reasonnable
if(nST<10)
Cov.Print();
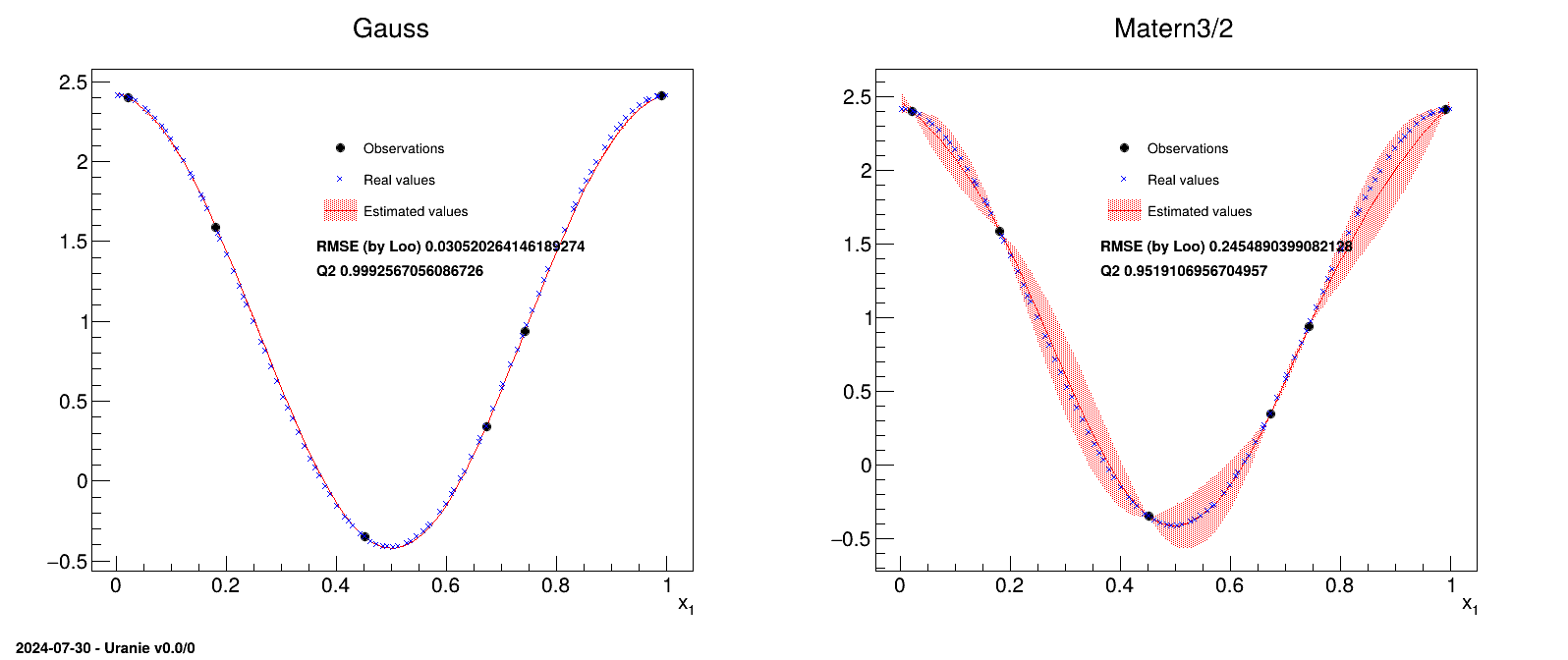
}The idea is to provide a simple example of a kriging usage, and an how to, to produce plots in one dimension to represent the results. This example is the one taken from [metho] that uses a very simple set of six points as training database:
#TITLE: utf-1D-train.dat #COLUMN_NAMES: x1|y 6.731290e-01 3.431918e-01 7.427596e-01 9.356860e-01 4.518467e-01 -3.499771e-01 2.215734e-02 2.400531e+00 9.915253e-01 2.412209e+00 1.815769e-01 1.589435e+00
The aim will be to get a kriging model that describes this dataset and to check its consistency over a certain number of points (here 100 points) which will be the testing database:
#TITLE: utf-1D-test.dat #COLUMN_NAMES: x1|y 5.469435e-02 2.331524e+00 3.803054e-01 -3.277316e-02 7.047152e-01 6.030177e-01 2.360045e-02 2.398694e+00 9.271965e-01 2.268814e+00 7.868263e-01 1.324318e+00 7.791920e-01 1.257942e+00 6.107965e-01 -8.514510e-02 1.362316e-01 1.926999e+00 5.709913e-01 -2.758435e-01 8.738804e-01 1.992941e+00 2.251602e-01 1.219826e+00 9.175259e-01 2.228545e+00 5.128368e-01 -4.096161e-01 7.692913e-01 1.170999e+00 7.394406e-01 9.062407e-01 5.364506e-01 -3.772856e-01 1.861864e-01 1.551961e+00 7.573444e-01 1.065237e+00 1.005755e-01 2.141109e+00 9.114685e-01 2.201001e+00 3.628465e-01 7.920271e-02 2.383583e-01 1.103353e+00 7.468092e-01 9.716492e-01 3.126209e-01 4.578112e-01 8.034716e-01 1.466248e+00 6.730402e-01 3.424931e-01 8.021345e-01 1.455015e+00 2.503736e-01 9.966807e-01 9.001793e-01 2.145059e+00 7.019990e-01 5.799112e-01 6.001746e-01 -1.432102e-01 4.925013e-01 -4.126441e-01 5.685795e-01 -2.849419e-01 1.238257e-01 2.007351e+00 2.825838e-01 7.124861e-01 4.025708e-01 -1.574002e-01 8.562999e-01 1.875879e+00 3.214125e-01 3.865241e-01 2.021767e-01 1.418581e+00 6.338581e-01 5.717657e-02 3.042007e-01 5.276410e-01 4.860707e-01 -4.088007e-01 9.645326e-01 2.379243e+00 3.583711e-02 2.378513e+00 2.143110e-01 1.314473e+00 7.299624e-01 8.224203e-01 2.719263e-02 2.393622e+00 3.321495e-01 3.020224e-01 8.642671e-01 1.930341e+00 8.893604e-01 2.086039e+00 1.119469e-01 2.078562e+00 9.859741e-01 2.408725e+00 5.594688e-01 -3.166326e-01 1.904448e-01 1.516930e+00 4.232618e-01 -2.529865e-01 1.402221e-01 1.899932e+00 2.647519e-01 8.691058e-01 1.667035e-01 1.706823e+00 2.332246e-01 1.148786e+00 8.324190e-01 1.700059e+00 4.743443e-01 -3.958790e-01 3.435927e-01 2.154677e-01 9.846049e-01 2.407603e+00 9.705327e-01 2.390043e+00 6.631883e-01 2.662970e-01 6.153726e-01 -5.862472e-02 4.632361e-01 -3.766509e-01 6.474053e-01 1.502050e-01 7.161034e-02 2.273461e+00 4.514511e-01 -3.489255e-01 5.976782e-02 2.315661e+00 8.361934e-01 1.729000e+00 5.280981e-01 -3.922313e-01 9.394759e-01 2.313181e+00 2.710088e-01 8.138628e-01 8.161943e-01 1.571375e+00 5.047683e-01 -4.135789e-01 8.427635e-02 2.220534e+00 3.540224e-01 1.400987e-01 4.698548e-03 2.413597e+00 9.124315e-02 2.188105e+00 9.996285e-01 2.414210e+00 4.167139e-01 -2.249546e-01 5.892062e-01 -1.978247e-01 2.929336e-01 6.231119e-01 4.456454e-01 -3.325379e-01 1.148699e-02 2.410532e+00 3.892636e-01 -8.548741e-02 7.188374e-01 7.248622e-01 3.697949e-01 3.323350e-02 6.864519e-01 4.502113e-01 1.586679e-01 1.767741e+00 6.603030e-01 2.445009e-01 6.277168e-01 1.721489e-02 4.305704e-01 -2.817686e-01 1.553435e-01 1.792379e+00 5.476842e-01 -3.512131e-01 8.475444e-01 1.813503e+00 9.527370e-01 2.352313e+00
In this example, two different correlation functions are tested and the obtained results are compared at the end.
void PrintSolutions(TDataServer *tdsObs, TDataServer *tdsEstim, string title)
{
const int nbObs = tdsObs->getNPatterns();
const int nbEst = tdsEstim->getNPatterns();
vector<double> Rxarr(nbEst,0), x_arr(nbEst,0), y_est(nbEst,0), y_rea(nbEst,0), std_var(nbEst,0);
tdsEstim->computeRank("x1");
tdsEstim->getTuple()->copyBranchData(&Rxarr[0],nbEst,"Rk_x1");
tdsEstim->getTuple()->copyBranchData(&x_arr[0],nbEst,"x1");
tdsEstim->getTuple()->copyBranchData(&y_est[0],nbEst,"yEstim");
tdsEstim->getTuple()->copyBranchData(&y_rea[0],nbEst,"y");
tdsEstim->getTuple()->copyBranchData(&std_var[0],nbEst,"vEstim");
vector<double> xarr(nbEst,0), yest(nbEst,0), yrea(nbEst,0), stdvar(nbEst,0);
int ind;
for (int i=0; i<nbEst; i++)
{
ind = int(Rxarr[i]) - 1;
xarr[ind] = x_arr[i];
yest[ind] = y_est[i];
yrea[ind] = y_rea[i];
stdvar[ind] = 2*sqrt(fabs(std_var[i]));
}
vector<double> xobs(nbObs,0); tdsObs->getTuple()->copyBranchData(&xobs[0],nbObs,"x1");
vector<double> yobs(nbObs,0); tdsObs->getTuple()->copyBranchData(&yobs[0],nbObs,"y");
TGraph *gobs = new TGraph(nbObs,&xobs[0],&yobs[0]);
gobs->SetMarkerColor(1); gobs->SetMarkerSize(1); gobs->SetMarkerStyle(8);
TGraphErrors *gest = new TGraphErrors(nbEst,&xarr[0],&yest[0],0,&stdvar[0]);
gest->SetLineColor(2); gest->SetLineWidth(1); gest->SetFillColor(2); gest->SetFillStyle(3002);
TGraph *grea = new TGraph(nbEst,&xarr[0],&yrea[0]);
grea->SetMarkerColor(4); grea->SetMarkerSize(1); grea->SetMarkerStyle(5); grea->SetTitle("Real Values");
TMultiGraph *mg = new TMultiGraph("toto", title.c_str());
mg->Add(gest,"l3"); mg->Add(gobs,"P"); mg->Add(grea,"P");
mg->Draw("A");
mg->GetXaxis()->SetTitle("x_{1}");
TLegend *leg= new TLegend(0.4,0.65,0.65,0.8);
leg->AddEntry(gobs, "Observations", "p");
leg->AddEntry(grea, "Real values", "p");
leg->AddEntry(gest, "Estimated values", "lf");
leg->Draw();
}
void modelerTestKriging()
{
//Create dataserver and read training database
TDataServer *tdsObs = new TDataServer("tdsObs","observations");
tdsObs->fileDataRead("utf-1D-train.dat");
const int nbObs = 6;
// Canvas, divided in 2
TCanvas *Can = new TCanvas("can","can",10,32,1600,700);
TPad *apad = new TPad("apad","apad",0, 0.03, 1, 1);
apad->Draw(); apad->Divide(2,1);
//Name of the plot and correlation functions used
string outplot="GaussAndMatern_1D_AllPoints.png";
string Correl[2] = {"Gauss","Matern3/2"};
//Pointer to needed objects, created in the loop
TGPBuilder *gpb[2];
TKriging *kg[2];
TDataServer *tdsEstim[2];
TLauncher2 *lkrig[2];
stringstream sstr;
//Looping over the two correlation function chosen
for (int imet=0; imet<2; imet++)
{
//Create the TGPBuiler object with chosen option and find optimal parameters
gpb[imet] = new TGPBuilder(tdsObs, "x1", "y", Correl[imet].c_str(), "");
gpb[imet]->findOptimalParameters("LOO", 100, "Subplexe", 1000);
//Get the kriging object
kg[imet] = gpb[imet]->buildGP();
//open the dataserver and read the testing basis
sstr.str(""); sstr << "tdsEstim_" << imet;
tdsEstim[imet] = new TDataServer(sstr.str().c_str(),"base de test");
tdsEstim[imet]->fileDataRead("utf-1D-test.dat");
//applied resulting kriging on test basis
lkrig[imet] = new TLauncher2(tdsEstim[imet], kg[imet], "x1", "yEstim:vEstim");
lkrig[imet]->solverLoop();
//do the plot
apad->cd(imet+1);
PrintSolutions(tdsObs, tdsEstim[imet], Correl[imet]);
stringstream sstr; TLatex *lat=new TLatex(); lat->SetNDC(); lat->SetTextSize(0.025);
sstr.str(""); sstr << "RMSE (by Loo) " << kg[imet]->getLooRMSE();
lat->DrawLatex(0.4,0.61,sstr.str().c_str());
sstr.str(""); sstr << "Q2 " << kg[imet]->getLooQ2();
lat->DrawLatex(0.4,0.57,sstr.str().c_str());
}
}
The first function of this macro (called PrintSolutions) is a complex part that will not
be detailed, used to represent the results.
The macro itself starts by reading the training database and storing it in a dataserver. A
TGPBuilder is created with the chosen correlation function and the hyper-parameters are
estimation by an optimisation procedure in:
gpb[imet] = new TGPBuilder(tdsObs, "x1", "y", Correl[imet].c_str(), "");
gpb[imet]->findOptimalParameters("LOO", 100, "Subplexe", 1000);
kg[imet] = gpb[imet]->buildGP()The last line shows how to build and retrieve the newly created kriging object.
Finally, this kriging model is tested against the training database, thanks to a TLauncher2
object, as following:
lkrig[imet] = new TLauncher2(tdsEstim[imet], kg[imet], "x1", "yEstim:vEstim");
lkrig[imet]->solverLoop();