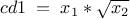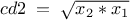Documentation
/ User's manual in Python
: 
This section describes the data import from an ASCII flat file in a TDataServer of Uranie
using the formalism of Salome tables and JSON format. In both cases, the ASCII file is composed of 2 parts: the header and the experiment matrix.
The header describes the information related to the database and related to its attributes:
the name of the database (optional);
the title of the database (optional);
the date of the database's saving (optional);
the name of variables (mandatory if the title/label's not specified). This information will enable to have access to theses variables either to transform them or to produce graphs. The chosen name has to be explicit but rather short (smaller than 50 characters);
the title/label of variables (mandatory if the name of variables is not specified). This information will be the axis tick marks when this variable is visualised on the plot. As it is possible to use LaTeX commands, the name (must be explicit and short) is distinguished from the label so as to obtain a good graph rendering.
the variable units (optional). This information aims at improving the graph rendering (the unit being displayed next to the label).
the nature of the variables (optional unless dealing with new types). This information is not mandatory if one wants to handle only double-precision variables. The possible value being "D" for double, "S" for string and "V" for vectors.
The second part is the numerical experiment matrix.
Using this formalism, it is enough to provide the name of attributes of the ASCII file in order to load the file in Uranie.
This is the main format used throughout the history of the Uranie-platform. The different header information are set thanks to keywords. A header line begins with the character '#' followed by a keyword characterising the type of information and by the ':' character . Then, the information are separated by the '|' character. The list of keywords is:
Table II.2. List of keywords of header in ASCII files.
| Keywords | Description |
|---|---|
| NAME | The name of the database |
| TITLE | The title of the database |
| DATE | The date of saving (only towards writing or export) |
| COLUMN_NAMES | The names of attributes |
| COLUMN_TYPES | The natures of attributes |
| COLUMN_TITLES | The titles of attributes |
| COLUMN_UNITS | The units of attributes |
An example is the file "geyser.dat"in the data directory of
installation of Uranie ( $URANIESYS/share/uranie/macros/geyser.dat)
> more $URANIESYS/share/uranie/macros/geyser.dat
#NAME: geyser
#TITLE: geyser data
#COLUMN_NAMES: x1| x2
#COLUMN_TITLES: x_{1}| "#delta P_{#sigma}"
#COLUMN_UNITS: Sec^{-1}| bar
3.600 79.000
1.800 54.000
3.333 74.000
2.283 62.000
4.533 85.000
2.883 55.000
4.700 88.000
3.600 85.000
1.950 51.000
4.350 85.000
1.833 54.000
3.917 84.000
4.200 78.000
1.750 47.000
4.700 83.000
2.167 52.000
... Uranie accepts several forms of file endings: it is possible the file ends with a white line or with a line with empty spaces, but also to end just after the last data.
Uranie does not accept data with "holes" (empty lines) like as follows in this modified version of the "geyser.dat" file:
#NAME: geyser
#TITLE: geyser data
#COLUMN_NAMES: x1| x2
#COLUMN_TITLES: x_{1}| "#delta P_{#sigma}"
#COLUMN_UNITS: Sec^{-1}| bar
3.600 79.000
1.800 54.000
3.333 74.000
2.283 62.000
4.533 85.000
2.883 55.000
4.700 88.000
3.600 85.000
1.950 51.000
4.350 85.000
1.833 54.000
3.917 84.000
4.200 78.000
1.750 47.000
4.700 83.000
2.167 52.000
...
In this case, Uranie considers that data processing ends with the white line located in the middle of the data lines. This would be equivalent to use the following data:
#NAME: geyser
#TITLE: geyser data
#COLUMN_NAMES: x1| x2
#COLUMN_TITLES: x_{1}| "#delta P_{#sigma}"
#COLUMN_UNITS: Sec^{-1}| bar
3.600 79.000
1.800 54.000
3.333 74.000
2.283 62.000
4.533 85.000Tip
Only the line associated to the keyword COLUMN_NAMES is mandatory except if COLUMN_TITLES is specified. Moreover, the keyword itself is also optional; the next line is correct # x1| x2 to specify both variables of the geyser data.
Warning
An empty line MUST be kept between the header and data matrix .
Particular case of strings and vector
The following example shows how to precise the content of vectors and strings if such information have to be
read. In this case, the field #COLUMN_TYPES: is mandatory and the way it works is
equivalent to the column name one (the delimiter is the "|" sign) but it needs only one letter to define the
type. Apart from that, the string can be written as it comes as long as it does not
contains blanks (!!), while the vectors values are dump with the same format as the double-precision
one, using a comma as separator. The following file is a correct input file for a TDataServer
#COLUMN_NAMES: day|place|hour|guest_list|food #COLUMN_TYPES: D|S|D|V|S 5 restaurant 4 2,3,4,5 chocolate 21 home 3 6,1,8,4,3 almond
The example shown above is working properly as there is no problematic behaviour in the data. Handling strings
and vectors can however be tricky as they respectively can be an empty string and an empty vector. This would
result in a missing number of field in a specific line which will make the
fileDataRead crashed. To avoid this, all the files used in the Launcher and Relauncher
(and the Salome-table discussed here as well) contains properties specific to both vectors and strings:
String properties: a character can be specified as begin and end for dumping and reading purpose. The ones chosen by default for the Salome-table format shown here being the double-quote sign ".
Vector properties: a character can be specified as begin, end and delimiter for dumping and reading purpose. The ones chosen by default for the Salome-table format shown here being respectively [, ] and the commas.
This results in the fact, that the following file gives the exact same dataserver as the one shown previously. It
is actually the style chosen when calling the exportData method of a TDataServer to allow the
user to handle empty strings and vectors if wanted.
#COLUMN_NAMES: day|place|hour|guest_list|food #COLUMN_TYPES: D|S|D|V|S 5 "restaurant" 4 [2,3,4,5] "chocolate" 21 "home" 3 [6,1,8,4,3] "almond"
Brought in version 3.9, the format has been implemented as it is broadly used to transmit data in a very simple way. A choice has been made on the way the header are displayed: a "_metadata" field is compulsory inside which the list of flag is gathered in Table II.3.
Table II.3. List of keywords of header in ASCII files.
| Keywords | Description |
|---|---|
| table_name | The name of the database |
| table_description | The title of the database |
| date | The date of saving (only towards writing or export) |
| short_names | The names of attributes |
| types | The natures of attributes |
| long_names | The titles of attributes |
| units | The units of attributes |
The second part that provides the data itself, looks alike a key-value table and handles easily all the attribute types. Here is an example of file with the "geyser" data content, shown previously:
{
"_metadata" :
{
"_comment" : "CurrentComment",
"date" : "Fri Oct 28 10:41:44 2016",
"short_names" : [ "x1", "x2", "geyser__n__iter__" ],
"table_description" : "Les donnees du geyser",
"table_name" : "geyser",
"types" : [ "D", "D", "D" ],
"units" : [ "Sec", "", "" ]
},
"items" :
[
{
"geyser__n__iter__" : 1.0,
"x1" : 3.60,
"x2" : 79.0
},
{
"geyser__n__iter__" : 2.0,
"x1" : 1.80,
"x2" : 54.0
},
{
"geyser__n__iter__" : 272.0,
"x1" : 4.4670,
"x2" : 74.0
}
]
}
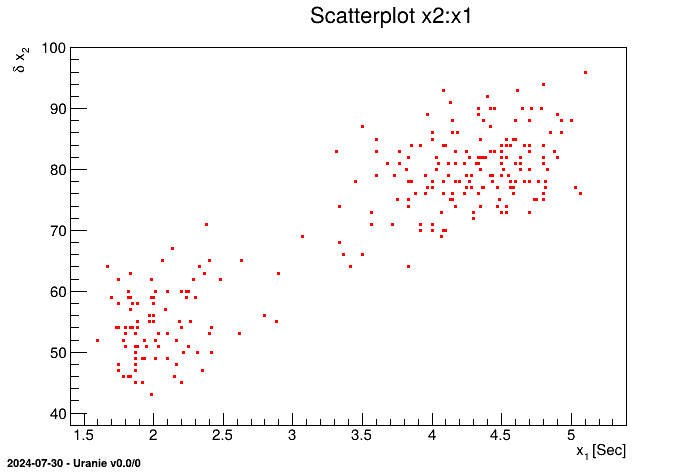
An example of import of the data file "geyser.dat" (available in the Uranie-macros folder) is
shown below leading to a 2D scatterplot of the variable x2 versus the variable
x1
from rootlogon import DataServer  tdsGeyser = DataServer.TDataServer("tdsgeyser", "Geyser database")
tdsGeyser = DataServer.TDataServer("tdsgeyser", "Geyser database")  tdsGeyser.fileDataRead("geyser.dat")
tdsGeyser.fileDataRead("geyser.dat") tdsGeyser.draw("x2:x1")
tdsGeyser.draw("x2:x1")
Description of the import of an ASCII file
Setting the namespace. This instruction is useless when the provided rootlogon has been loaded as all Uranie-namespaces have been loaded as well. | |
Defining a pointer tdsGeyser to an object of type | |
Loading data contained in an ASCII file | |
Plot of the scatterplot of the variable x2 versus the variable x1 |
The obtained graph is the following:
Various examples of macros loading data in a TDataServer with different treatment applied on, are provided in the use-case
chapter of this user manual, between Section XIV.3.3 and Section XIV.3.7.
From a TTree object (or any of its derived-object) contained in a ROOT-file, it is possible to import data, with or
without selection of a variable and addition of other ones through formula then deletions of patterns ensuring a
criterion
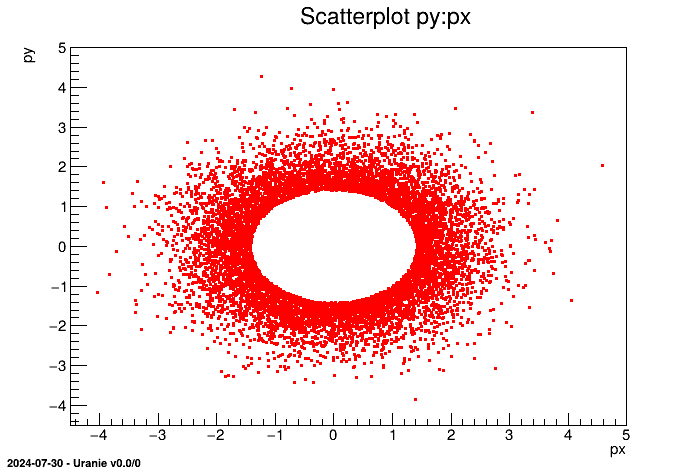
from rootlogon import DataServer  tds = DataServer.TDataServer()
tds = DataServer.TDataServer()  tds.ntupleDataRead("hsimple.root","ntuple","px*px:*:py*py","px*px+py*py<2.0")
tds.ntupleDataRead("hsimple.root","ntuple","px*px:*:py*py","px*px+py*py<2.0")  tds.draw("py:px")
tds.draw("py:px") 
Description of data importation of a TTree from a ROOT file
Specification of the namespace. | |
Creation of a pointer tds to an object of type | |
Fill the | |
Plots scatterplot of the variable py versus the variable px |
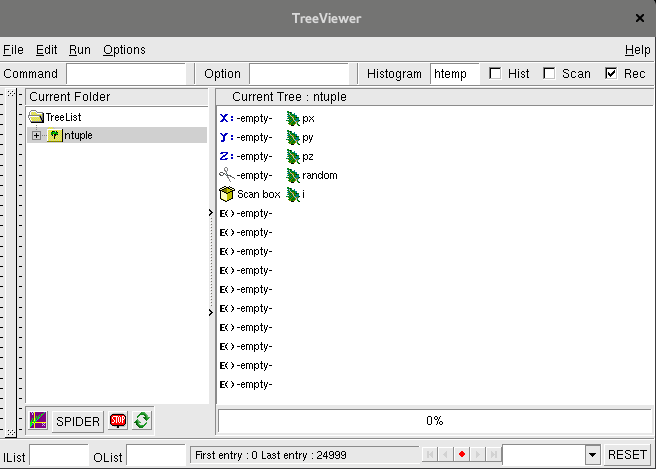
In this case, tds is constructed from the TTree
ntuple contained in the file hsimple.root where all initial variables are
kept (* character). Figure II.37 shows the content of the
ntuple. Two new variables are then added on top, defined by the equations "px*py" and "py*px" on
both side of the "*" string. A cut is finally done, to exclude all data that would satisfy the following equation
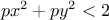
The obtained graph is as follow:
Attributes can always be added to an existing TDataServer object, whether it is empty (just after its
constructor) or not (after the data loading from either an ASCII file, or a TTree or a database
of type SQL). A simple example is provided and decomposed in Section XIV.3.1.
First of all, let us consider the case of an empty TDataServer. We add attributes using the method
addAttribute( TAttribute *att).
tds = DataServer.TDataServer("tds", "new TDataServer")
tds.addAttribute(DataServer.TAttribute("x1")) 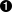 tds.addAttribute(DataServer.TAttribute("x2", 2.5, 5.))
tds.addAttribute(DataServer.TAttribute("x2", 2.5, 5.)) 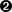 tds.addAttribute( "x3" )
tds.addAttribute( "x3" )  tds.addAttribute( "x4", 2.5, 5.)
tds.addAttribute( "x4", 2.5, 5.) 
Description of attributes adding to an empty TDataServer
Adding a new attribute x1 to the TDataServer from a
| |
Adding a new attribute x2 to the TDataServer from a
| |
Equivalent to previous one: adding a new attribute x3 to the TDataServer just by giving its name (minimal constructor). | |
Equivalent to previous one: adding a new attribute x4 to the TDataServer by giving its name and the extreme values (minimal and maximal). |
The difference between the methods with new in it and the others, is basically arising from the way one
handles the memory. The last ones (3 and 4) allow the user not to worry about anything, while, in the case of
implementation 1 and 2, one should be aware that every new should imply at some point a
delete. For most user, this is not of utmost importance as usual scripts would contain very few
new command and no loop. If this is not the case (for instance if one does have loop and many object
creation in it) do not hesitate to contact the Uranie-team to prevent any slowing down of the code.
The specification of a TAttribute is further detailed in Section II.2.
We can define new attributes using mathematical expressions with respect to other existing attributes. The name and the mathematical expression are the only mandatory arguments; its title and unit can also be precised, but both arguments are optional.
tdsGeyser = DataServer.TDataServer("tdsgeyser", "Geyser DataSet")
tdsGeyser.fileDataRead("geyser.dat")
tdsGeyser.addAttribute("cd1","sqrt(x2) * x1") 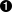 tdsGeyser.addAttribute("cd2","sqrt(x2*x1)","#Delta p_{#sigma}","sec^{-1}")
tdsGeyser.addAttribute("cd2","sqrt(x2*x1)","#Delta p_{#sigma}","sec^{-1}")  tdsGeyser.draw("cd2:cd1")
tdsGeyser.draw("cd2:cd1") 
Description attribute adding to a TDataServer from formulas
Adding a new attribute cd1 to the TDataServer defined by a mathematical expression as a function of x1 and x2 attributes:
| |
Adding a new attribute cd2 to the TDataServer with a mathematical formula, precising its title and unit:
| |
Plots the scatterplot of the variable cd2 versus the variable cd1 |
The obtained graph is:
This operation is available with vector-type attribute as well. The results depends on the nature of the
attributes involved in the formula, their content and the nature of the operation. As an example, a simple dataserver
is created from the dummy file tdstest.dat:
#COLUMN_NAMES: x| y| a| v #COLUMN_TYPES: V|V|D|V 1,2,3 4,5,6 2 1,2,3 7,8,9 1,2 4 4,5,6 1,4,8 2,5,4 5 7,8,9
It contains two vectors whose size are not constant and a double. The four usual operations have been performed (addition, subtraction, multiplication and division) using the double and a vector but also using the two vectors. The code is shown here:
"""
Example of attribute handling
"""
from rootlogon import DataServer
tdsop = DataServer.TDataServer("foo", "poet")
tdsop.fileDataRead("tdstest.dat")
tdsop.addAttribute("x*y", "x*y") # Multiply two vectors
tdsop.addAttribute("xovy", "x/y") # Divide two vectors
tdsop.addAttribute("x-y", "x-y") # Subtract two vectors
tdsop.addAttribute("x+y", "x+y") # Add two vectors
tdsop.addAttribute("x*a", "x*a") # Multiply a vector and a double
tdsop.addAttribute("xova", "x/a") # Divide a vector and a double
tdsop.addAttribute("x-a", "x-a") # Subtract a vector and a double
tdsop.addAttribute("x+a", "x+a") # Add a vector and a double
tdsop.scan("x:y:a:x*y:xovy:x+y:x-y:x*a:xova:x+a:x-a", "",
"colsize=3 col=1:1:1::4::::4::")
and it gives as a results:
************************************************************************************* * Row * Instance * x * y * a * x*y * xovy * x+y * x-y * x*a * xova * x+a * x-a * ************************************************************************************* * 0 * 0 * 1 * 4 * 2 * 4 * 0.25 * 5 * -3 * 2 * 0.5 * 3 * -1 * * 0 * 1 * 2 * 5 * 2 * 10 * 0.4 * 7 * -3 * 4 * 1 * 4 * 0 * * 0 * 2 * 3 * 6 * 2 * 18 * 0.5 * 9 * -3 * 6 * 1.5 * 5 * 1 * * 1 * 0 * 7 * 1 * 4 * 7 * 7 * 8 * 6 * 28 * 1.75 * 11 * 3 * * 1 * 1 * 8 * 2 * 4 * 16 * 4 * 10 * 6 * 32 * 2 * 12 * 4 * * 1 * 2 * 9 * * 4 * 0 * * 0 * 0 * 36 * 2.25 * 13 * 5 * * 2 * 0 * 1 * 2 * 5 * 2 * 0.5 * 3 * -1 * 5 * 0.2 * 6 * -4 * * 2 * 1 * 4 * 5 * 5 * 20 * 0.8 * 9 * -1 * 20 * 0.8 * 9 * -1 * * 2 * 2 * 8 * 4 * 5 * 32 * 2 * 12 * 4 * 40 * 1.6 * 13 * 3 * *************************************************************************************
Warning
This section is discussing the merging of two TDataServer not their concatenation. The first operation consists in adding
new attributes from an existing TDataServer into another existing one, while the seconds consists in adding the content
of two TTree object with the exact same structure. For the merging operation, a specific
method TDataServer::merge has been written, while for the concatenation, the interested user is
invited to look at the TChain::Merge method from ROOT.
It is sometimes necessary to merge two TDataServer to form a single one. Since the
merging is done line by line, one has to check that both objects contain the same number of patterns. In
Uranie-version older than 3.10.0 it was assumed that the patterns were exactly
stored in the same order. Now the method is looking for the iterator of both TDataServer objects and it checks that both
iterators contain the same value all along (not necessary in the same order, for instance when dealing with
distributed computations). If the iterators are not found or if some iterator's values are found in one iterator but
not the other (possible in some rare cases such as OAT sampling), the merging is done
line-by-line and a warning is displayed.
This operation is common when you want to build a surface response between output variables Y and predictors X and these data are located in two different files.
Warning
The 2 objects must have the same number of patterns.
Let's take a simple example. Assuming that we have 2 TDataServer tds1 and tds2 respectively located in the ASCII files tds1.dat and
tds2.dat. Another example is also provided in Section XIV.3.2.
Data file tds1.dat
#COLUMN_NAMES: x| dy| z| theta
#COLUMN_TITLES: x_{n}| "#delta y"| ""| #theta
#COLUMN_UNITS: N| Sec| KM/Sec| M^{2}
1 1 11 11
1 2 12 21
1 3 13 31
2 1 21 12
2 2 22 22
2 3 23 32
3 1 31 13
3 2 32 23
3 3 33 33
Data file tds2.dat
#COLUMN_NAMES: x2| y| u| ua 1 1 102 11 1 2 104 12 1 3 106 13 2 1 202 21 2 2 204 22 2 3 206 23 3 1 302 31 3 2 304 32 3 3 306 33
Both TDataServer incorporate 9 patterns.
These 2 ASCII files must be loaded in 2 TDataServer (cf Section II.3.2), the merging being done by calling the
merge method of the first TDataServer
"""
Example of data merging (merging two files)
"""
from rootlogon import DataServer
tds1 = DataServer.TDataServer()
tds1.fileDataRead("tds1.dat")
tds2 = DataServer.TDataServer()
tds2.fileDataRead("tds2.dat")
tds1.merge(tds2)
Thus, the object tds1 also contains the attributes of the second TDataServer
tds2
************************************************************** * Row * tds * x. * dy * z. * theta * x2 * y. * u.u * ua * ************************************************************** * 0 * 1 * 1 * 1 * 11 * 11 * 1 * 1 * 102 * 11 * * 1 * 2 * 1 * 2 * 12 * 21 * 1 * 2 * 104 * 12 * * 2 * 3 * 1 * 3 * 13 * 31 * 1 * 3 * 106 * 13 * * 3 * 4 * 2 * 1 * 21 * 12 * 2 * 1 * 202 * 21 * * 4 * 5 * 2 * 2 * 22 * 22 * 2 * 2 * 204 * 22 * * 5 * 6 * 2 * 3 * 23 * 32 * 2 * 3 * 206 * 23 * * 6 * 7 * 3 * 1 * 31 * 13 * 3 * 1 * 302 * 31 * * 7 * 8 * 3 * 2 * 32 * 23 * 3 * 2 * 304 * 32 * * 8 * 9 * 3 * 3 * 33 * 33 * 3 * 3 * 306 * 33 * **************************************************************
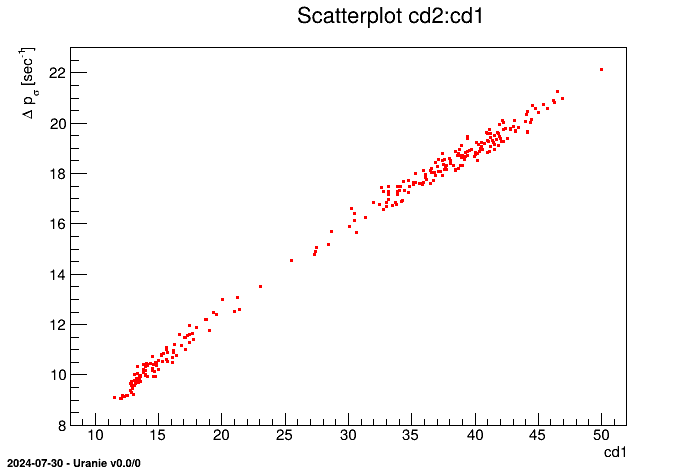
It can be necessary during a study to apply filters on the patterns; i.e. to include or to exclude patterns depending
on criterion. For example, to select the patterns with x1 lower than 3.0 and x2 lower than 55.0 from the
geyser database, Uranie code is as follows:
tdsGeyser.setSelect("( x1<3.0 ) && ( x2<55.)")
tdsGeyser.draw("x2:x1") The obtained figure is:
The result of the scan method applied on this TDataServer object yields:
************************************************ * Row * x1 * x2 * n__iter__ * ************************************************ * 1 * 1.8 * 54 * 2 * * 8 * 1.95 * 51 * 9 * * 10 * 1.833 * 54 * 11 * * 13 * 1.75 * 47 * 14 * * 15 * 2.167 * 52 * 16 * ... * 268 * 2.15 * 46 * 269 * * 270 * 1.817 * 46 * 271 * ************************************************ ==> 53 selected entries
We have obtained 53 patterns among 278 respecting the given criterion without having to specify this criterion for
the draw and scan calls. To get the same result, we could have
executed the following command as well:
tdsGeyser.draw("x2:x1", "( x1<3.0 ) && ( x2<55.)")
# tdsGeyser.scan("*", "( x1<3.0 ) && ( x2<55.)") However, in this case, we have to repeat the criterion for each command.
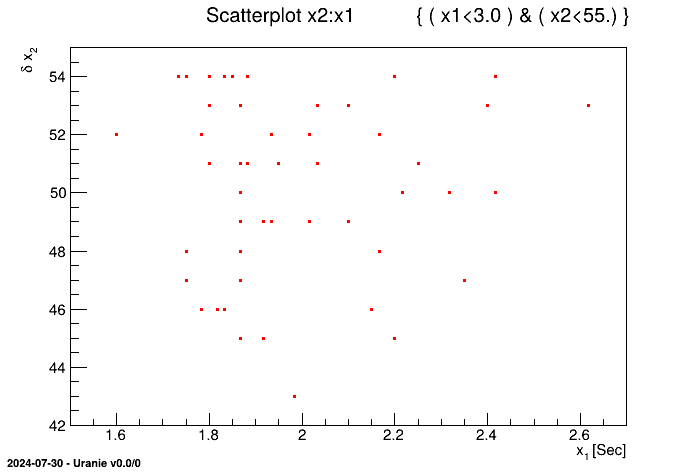
It is also possible to exclude patterns coming from TDataServer with the
setCut method.
tdsGeyser.setCut("x1 >= 3.")
tdsGeyser.draw("x2:x1") The obtained figure is as follows:
It can be noticed in the title of Figure II.41, that it simply corresponds to
the opposite of criterion's meaning with respect to the one given by the setCut method with
the ! ( ) character. Thus, the setCut method consists in
passing on the negation of the criterion to the setSelect command.
Finally, it is perfectly possible to delete the current filters with the methods clearSelect
and clearCut, and retrieving the unfiltered results.
Tip
Every modifications of the ongoing selection (meaning doing a new selection or removing it) is now clearing automatically the vectors that contain statistical properties of attributes and the database of already computed quantiles. This is reminded with an information line shown below:
<URANIE::INFO> Selection is changing ==> clearing the TAttribute computed statistics and quantiles
In the same way as the data are imported from an ASCII file, we can also save the data of a TDataServer in an ASCII
file. Currently, four methods of export are available in Uranie:
using the same format as that observed during import ("Salome Table");
using a C file containing the data vectors that can be inserted in a C program.
using the NeMo format: the generated file is useful for the NeMo tool for constructing neural response surface developed at STMF.
using the JSON format: the generated file is easily transferable to any other program that include the JSON protocol. This file can also be read through the
fileDataReadJSonmethod of aTDataServerobject.
tdsGeyser = DataServer.TDataServer("tdsgeyser", "geyser database")
tdsGeyser.fileDataRead("geyser.dat")
tdsGeyser.addAttribute("y", "sqrt(x2) * x1")
tdsGeyser.exportData("newfile.dat")  tdsGeyser.exportDataHeader("newfile.C", "x1:x2:y")
tdsGeyser.exportDataHeader("newfile.C", "x1:x2:y")  tdsGeyser.exportDataNeMo("newfile.nemo", "x1:x2", "y", "x2<75.0")
tdsGeyser.exportDataNeMo("newfile.nemo", "x1:x2", "y", "x2<75.0")  tdsGeyser.exportDataJSon("newfile.json")
tdsGeyser.exportDataJSon("newfile.json") 
Data export from a TDataServer in an ASCII file
Export the data of the #NAME: tdsgeyser #TITLE: Database of the geyser #DATE: Tue Oct 9 15:41:29 2007 #COLUMN_NAMES: x1| x2| y| n__iter__ 3.600000000e+00 7.900000000e+01 3.199749990e+01 1 1.800000000e+00 5.400000000e+01 1.322724461e+01 2 3.333000000e+00 7.400000000e+01 2.867155012e+01 3 2.283000000e+00 6.200000000e+01 1.797635998e+01 4 4.533000000e+00 8.500000000e+01 4.179219502e+01 5 ... 2.150000000e+00 4.600000000e+01 1.458200946e+01 269 4.417000000e+00 9.000000000e+01 4.190334127e+01 270 1.817000000e+00 4.600000000e+01 1.232349358e+01 271 4.467000000e+00 7.400000000e+01 3.842658697e+01 272 | |
Exports the data of attributes x1, x2 and y of the // File "newfile.C" generated by ROOT v5.17/04
// DateTime Tue Oct 9 15:41:30 2007
// DataServer: Name="tdsgeyser" Title="Database of the geyser" Select=""
#define essai_nPattern 272
// Attribute Name="x1" Title=" x_{1}"
Double_t x1[essai_nPattern] = {
3.600000000e+00,
1.800000000e+00,
...
1.817000000e+00,
4.467000000e+00,
};
// End of attribute x1
// Attribute Name="x2" Title=" #delta x_{2}"
Double_t x2[essai_nPattern] = {
7.900000000e+01,
5.400000000e+01,
...
1.232349358e+01,
3.842658697e+01,
};
// End of attribute y
// End of File newfile.C
| |
Exports the data of the #NombreExemples 126 #NombreEntrees 2 #NombreSorties 1 1.800000000e+00 5.400000000e+01 1.322724461e+01 3.333000000e+00 7.400000000e+01 2.867155012e+01 2.283000000e+00 6.200000000e+01 1.797635998e+01 2.883000000e+00 5.500000000e+01 2.138090024e+01 ... 2.150000000e+00 4.600000000e+01 1.458200946e+01 1.817000000e+00 4.600000000e+01 1.232349358e+01 4.467000000e+00 7.400000000e+01 3.842658697e+01 | |
Exports the data of the
{
"_metadata" :
{
"_comment" : "CurrentComment",
"date" : "Fri Oct 28 10:41:44 2016",
"short_names" : [ "x1", "x2", "geyser__n__iter__" ],
"table_description" : "Les donnees du geyser",
"table_name" : "geyser",
"types" : [ "D", "D", "D" ],
"units" : [ "Sec", "", "" ]
},
"items" :
[
{
"geyser__n__iter__" : 1.0,
"x1" : 3.60,
"x2" : 79.0
},
{
"geyser__n__iter__" : 2.0,
"x1" : 1.80,
"x2" : 54.0
},
{
"geyser__n__iter__" : 272.0,
"x1" : 4.4670,
"x2" : 74.0
}
]
}
|