Documentation
/ Manuel utilisateur en Python
: 
The idea of the One factor At a Time (OAT) design-of-experiments is to observe the evolution of phenomena when one input factor is modified while all the others remain unchanged.
This design-of-experiments can be used to compute partial derivatives or sensitivities. For the latter application, it is interesting as it provides a very simple and inexpensive way to evaluate the sensitivity of a phenomena to its input factors. However, it might not be the best solution as it does not explore multi-factorial and non-linear effects.
In Uranie, each factor of the OAT design takes at least three values: a nominal value, a
lower value (smaller than the nominal one) and an upper value (greater than
the nominal one). When a factor is not "modified" it is set to its nominal value. This leads to a design of
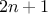 experiments, where
experiments, where
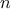 is the number of factors, and
is the number of factors, and
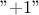 refers to a reference experiment,
where no factor is modified.
refers to a reference experiment,
where no factor is modified.
Two classes can produce OAT designs:
TOATSampling: simple, but limited in its functionality. It is used by some classes of Uranie.TOATDesign: an improved version of the previous one, more user oriented.
As TOATSampling will become deprecated in a future version
of Uranie the present document will focus on the usage of TOATDesign.
The easiest way to create an OAT design-of-experiments using TOATDesign is to proceed as follows:
create a dataserver with a list of attributes corresponding to the input factors;
set the default value of each attribute to the nominal value;
create a
TOATDesignobject, with the dataserver as first parameter;define the maximum range of variation of the factors using the
setRangefunction;generate the OAT design-of-experiments using the
generateSamplefunction.
At this point, the dataserver contains an OAT design-of-experiments where each factor of interest is modified twice. Below is an example:
"""
Example of simple OAT DoE
"""
from rootlogon import DataServer, Sampler
# step 1
tds = DataServer.TDataServer("tdsoat", "Dataserver simple OAT design")
tds.addAttribute(DataServer.TAttribute("x1"))
tds.addAttribute(DataServer.TAttribute("x2"))
# step 2
tds.getAttribute("x1").setDefaultValue(0.0)
tds.getAttribute("x2").setDefaultValue(10.0)
# step 3
oatSampler = Sampler.TOATDesign(tds)
# step 4
use_percentage = True
oatSampler.setRange("x1", 2.0)
oatSampler.setRange("x2", 40.0, use_percentage)
# step 5
oatSampler.generateSample()
# display
tds.scan()
This example produces:
**************************************************************************** * Row * tdsoat__n__iter * x1 * x2 * __nominal_set__ * __modified_att_ * **************************************************************************** * 0 * 1 * 0 * 10 * 1 * -1 * * 1 * 2 * -1 * 10 * 1 * 1 * * 2 * 3 * 1 * 10 * 1 * 1 * * 3 * 4 * 0 * 8 * 1 * 2 * * 4 * 5 * 0 * 12 * 1 * 2 * ****************************************************************************
In the example above, we have two input factors: x1 and x2. Their nominal values are respectively 0.0 and 10.0, and their maximum variation ranges are 2.0 and 40% of 10.0, i.e. 4.0.
Tip
To indicate that the range of "x2" is a percentage of its nominal value, we simply need to set the third parameter of the setRange function to TRUE. This parameter is set to FALSE by default, which means that the value of the range is considered to be "absolute".
The generated OAT design thus contains 5 experiments:
the reference, where x1 and x2 are set to their nominal values;
two variations of x1, where it equals -1.0 and 1.0 while x2 remains equal to 10.0;
two variations of x2, where it equals 8.0 and 12.0 while x1 is set back to 0.0;
It also contains two new attributes, automatically added by Uranie:
__nominal_set__: identifies which set of nominal values is used as a reference. In this case, we have only one set, thus __nominal_set__'s value remains equal to 1. This is further discussed in Section III.6.3.5
__modified_att__: identifies which factor has been modified in the current experiment. The value is the index of the corresponding attribute in the dataserver. A value equal to -1 means that all the factors have their nominal values.
Warning
The index of an attribute in the dataserver can be different from the one printed by the scan function, or inside an output file. To be certain to retrieve the correct number, always use the
TDataServer::getAttributeIndexfunction.
When creating a new OAT sampler object, the following options are available:
sampling mode: determines how the modified factor's new values are selected inside the interval
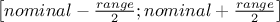 . It can be either:
. It can be either:
regular: an even number of new values and the nominal value are regularly distributed along the interval.
lhs, srs or random: the new values are randomly chosen inside the interval using a Latin Hypercube Sampling ("lhs") or a Standard Random Sampling ("srs" and "random") (cf. Figure III.3). The sample's distribution is given by the type of the attribute representing the factor (cf. Section II.2.5).
number of modifications: the number of new values taken by a modified factor. It must be greater or equal to 2. If the sampling mode is "regular" and the given number
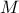 is odd, the
actual number of modifications will be
is odd, the
actual number of modifications will be 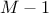 .
.
In the example of Section III.6.3.1, we used the default options for the OAT sampler, namely:
sampling mode: regular;
number of modifications: 2.
In the next examples, we present the results of choosing different options.
In the example of Section III.6.3.1, if we change the "step 3" to:
oatSampler = Sampler.TOATDesign(tds, "regular", 4)the OAT design becomes:
******************************************************************************* * Row * tdsoat__n__iter * x1.x * x2. * __nominal_set__ * __modified_att_ * ******************************************************************************* * 0 * 1 * 0 * 10 * 1 * -1 * * 1 * 2 * -1 * 10 * 1 * 1 * * 2 * 3 * 1 * 10 * 1 * 1 * * 3 * 4 * -0.5 * 10 * 1 * 1 * * 4 * 5 * 0.5 * 10 * 1 * 1 * * 5 * 6 * 0 * 8 * 1 * 2 * * 6 * 7 * 0 * 12 * 1 * 2 * * 7 * 8 * 0 * 9 * 1 * 2 * * 8 * 9 * 0 * 11 * 1 * 2 * *******************************************************************************
Each factor is modified 4 times, and its values are regularly spaced over the interval of variation (see Section XIV.4.6 for complete code).
In order to have randomly distributed values over the interval, the example's code needs further modifications.
To produce a random sampling, the attributes representing the factors must belong to the TStochasticAttribute family (cf. Section II.2.5). We thus need to modify the "step 1" of example of Section III.6.3.1 to:
# step 1
tds = DataServer.TDataServer("tds", "Data server for simple OAT design")
tds.addAttribute(DataServer.TUniformDistribution("x1", -5.0, 5.0))
tds.addAttribute(DataServer.TNormalDistribution("x2", 11.0, 1.0))
Tip
There is no a priori relationship between the distribution of the attribute and the nominal value and range of the factor it represents. However, a good practice is to insure that the probability density over the whole factor's range is never null.
The "step 2" of the example of Section III.6.3.1 does not need to be modified. The "step 3", on the other hand, becomes:
# step 3
oatSampler = Sampler.TOATDesign(tds, "lhs", 1000)By choosing the "lhs" mode, we ask for a random sampling over the range of the factor (defined in "step 4"). Here, we also ask for 1000 [2] modifications of each factor.
The rest of the script remains unchanged. We modify the "//display" section in order to visualise histograms of the sampling, instead of a long list of numbers:
# display
c = ROOT.TCanvas("can")
c.Divide(2,1)
c.cd(1)
tds.draw("x1", "__modified_att__ == 1")
c.cd(2)
tds.draw("x2", "__modified_att__ == 2")
Tip
The two calls to draw are an illustration of the use of the "__modified_att__"
attribute. Here, it allows to filter out the data, by keeping only the experiments where the interesting factor
is modified.
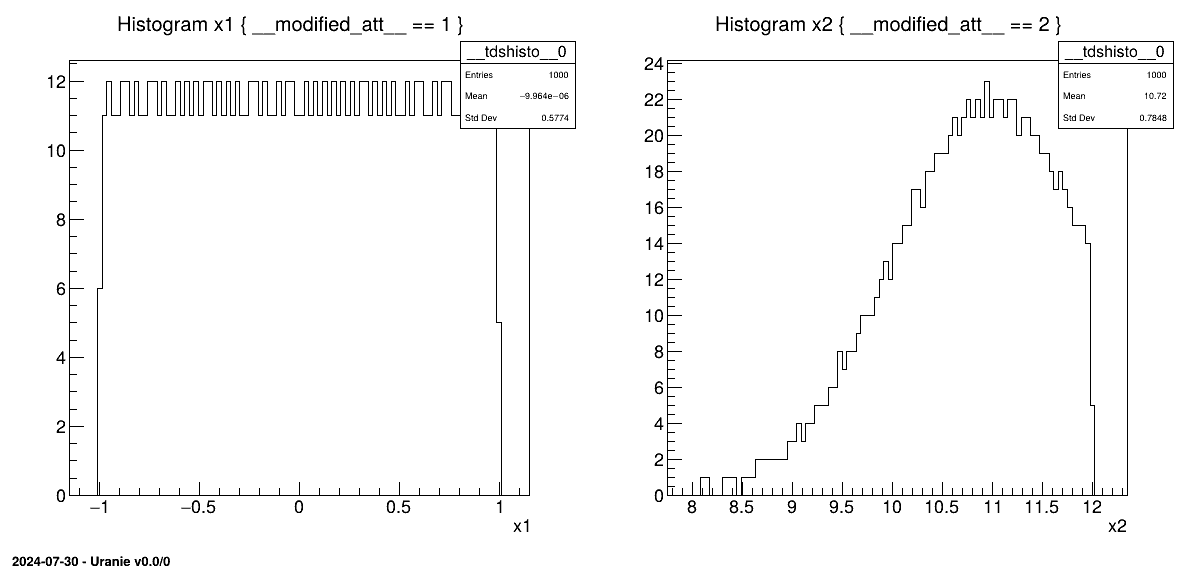
The resulting histograms are shown in figure Figure III.15. The left histogram shows the distribution of data for the "x1" attribute (uniform distribution) and the right, for the "x2" attribute (gaussian distribution). The latter shows how the gaussian distribution is truncated by the choice of the nominal value and the range (the complete code can be found in Section XIV.4.7).
If the factors have more than one possible nominal value, the OAT sampler can automatically build a design-of-experiments for each set of nominal values stored in the dataserver.
This information can be set manually, but an easier way is to write it inside a "Salome table" file (cf. Section II.3 for a description of the format). Below is a simple example of such a file:
#FILE_NAME: myNominalValues.dat
#COLUMN_NAMES: x1 | x2
0.0 10.0
5.0 3.0
-5.0 17.0
Still starting from the script of the example of Section III.6.3.1, the only step requiring modifications is the first one:
# step 1
tds = DataServer.TDataServer("tds", "Data server for simple OAT design")
tds.fileDataRead("myNominalValues.dat")The "step 2" is now useless and must be removed. The nominal values of the factors will be automatically loaded from the dataserver.
Now if we run the modified script, the result is:
******************************************************************************** * Row * tdsoat__n__iter * x1 * x2 * __nominal_set__ * __modified_att_ * ******************************************************************************** * 0 * 1 * 0 * 10 * 1 * -1 * * 1 * 2 * -1 * 10 * 1 * 0 * * 2 * 3 * 1 * 10 * 1 * 0 * * 3 * 4 * 0 * 8 * 1 * 1 * * 4 * 5 * 0 * 12 * 1 * 1 * * 5 * 6 * 5 * 3 * 2 * -1 * * 6 * 7 * 4 * 3 * 2 * 0 * * 7 * 8 * 6 * 3 * 2 * 0 * * 8 * 9 * 5 * 2.4 * 2 * 1 * * 9 * 10 * 5 * 3.6 * 2 * 1 * * 10 * 11 * -5 * 17 * 3 * -1 * * 11 * 12 * -6 * 17 * 3 * 0 * * 12 * 13 * -4 * 17 * 3 * 0 * * 13 * 14 * -5 * 13.6 * 3 * 1 * * 14 * 15 * -5 * 20.4 * 3 * 1 * ********************************************************************************
The generated design contains 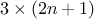 experiments. The attribute "__nominal_set__" now varies from 1
to 3, indicating which set of nominal value is taken as reference. The complete code can be found in Section XIV.4.8.
experiments. The attribute "__nominal_set__" now varies from 1
to 3, indicating which set of nominal value is taken as reference. The complete code can be found in Section XIV.4.8.
We have seen that it is possible to interpret the range of variation as a percentage of the nominal value. This is often enough to adapt the range to very different nominal values. However, in some contexts it can be useful to really modify the range of a factor.
This information can also be read from a data file. For example, our previous file can be modified to add a new attribute representing the range of one of the factors:
#FILE_NAME: myNominalValues.dat #COLUMN_NAMES: x1 | x2 | rx1 0.0 10.0 2.0 5.0 3.0 0.4 -5.0 17.0 6.0
Tip
The name of the attribute and the order in which it appears in the file is meaningless. Actually, any attribute can be considered as a range.
The modifications of "step 1" and the removal of "step 2" in the example of Section III.6.3.5 are still valid, while "step 4" must be modified in order to tell the OAT sampler that the range of "x1" is represented by another attribute:
# step 4
use_percentage = True
oatSampler.setRange("x1", "rx1")
oatSampler.setRange("x2", 40.0, use_percentage)Now, for each new set of nominal values, the value of "rx1" will become the range of "x1".
The result of the modified script (which can be found in Section XIV.4.8) is:
************************************************************************************ * Row * tds__n__iter * x1 * x2 * rx1 * __nominal_set__ * __modified_att_ * ************************************************************************************ * 0 * 1 * 0 * 10 * 2 * 1 * -1 * * 1 * 2 * -1 * 10 * 2 * 1 * 0 * * 2 * 3 * 1 * 10 * 2 * 1 * 0 * * 3 * 4 * 0 * 8 * 2 * 1 * 1 * * 4 * 5 * 0 * 12 * 2 * 1 * 1 * * 5 * 6 * 5 * 3 * 0.4 * 2 * -1 * * 6 * 7 * 4.8 * 3 * 0.4 * 2 * 0 * * 7 * 8 * 5.2 * 3 * 0.4 * 2 * 0 * * 8 * 9 * 5 * 2.4 * 0.4 * 2 * 1 * * 9 * 10 * 5 * 3.6 * 0.4 * 2 * 1 * * 10 * 11 * -5 * 17 * 6 * 3 * -1 * * 11 * 12 * -8 * 17 * 6 * 3 * 0 * * 12 * 13 * -2 * 17 * 6 * 3 * 0 * * 13 * 14 * -5 * 13.6 * 6 * 3 * 1 * * 14 * 15 * -5 * 20.4 * 6 * 3 * 1 * ************************************************************************************
If
we compare this design to the previous one, we can see that the column for "x2" is unchanged, while "x1" is
modified according to the value of "rx1". We can also note that "rx1" is never modified (in the OAT way). This is
because no call to the function setRange was done for it. Only the
attributes with an associated range are modified by the sampling procedure.
To finish this description of the OAT sampler of Uranie, here are some general remarks to answer frequently asked questions or to inform the user about the evolution of the class.
It is not possible to use random modes when data are loaded from a file.
The value of a range can always be interpreted as percentage of the nominal value, even if the range is read from the dataserver. Please refer to the developer documentation of the function
URANIE::Sampler::TOATDesign::setRangefor details.
[2] This is a ridiculously high number for an OAT design whose aim is, precisely, to provide a small and simple design-of-experiments. We do this only to be able to visualise nice histograms !